Login
Registration enables users to use special features of this website, such as past
order histories, retained contact details for faster checkout, review submissions, and special promotions.
order histories, retained contact details for faster checkout, review submissions, and special promotions.
Forgot password?
Registration enables users to use special features of this website, such as past
order histories, retained contact details for faster checkout, review submissions, and special promotions.
order histories, retained contact details for faster checkout, review submissions, and special promotions.
Quick Order
Products
Antibodies
ELISA and Assay Kits
Research Areas
Infectious Disease
Resources
Purchasing
Reference Material
Contact Us
Location
Corporate Headquarters
Vector Laboratories, Inc.
6737 Mowry Ave
Newark, CA 94560
United States
Telephone Numbers
Customer Service: (800) 227-6666 / (650) 697-3600
Contact Us
Additional Contact Details
Login
Registration enables users to use special features of this website, such as past
order histories, retained contact details for faster checkout, review submissions, and special promotions.
order histories, retained contact details for faster checkout, review submissions, and special promotions.
Forgot password?
Registration enables users to use special features of this website, such as past
order histories, retained contact details for faster checkout, review submissions, and special promotions.
order histories, retained contact details for faster checkout, review submissions, and special promotions.
Quick Order
Colorectal Cancer Immunohistochemistry Markers
Colorectal carcinoma (CRC) affects 4% of the population, and up to 30% of patients have a family history of the disease. Identified hereditary syndromes such as Lynch syndrome (Hereditary
nonpolyposis colorectal cancer or HNPCC) and Familial adenomatous polyposis (FAP), however, only account for 10% of patients (Liccardo, 2017) and risk factors such as age, diet and smoking,
or diseases such as diabetes and chronic inflammatory bowel diseases have also been implicated as increasing risk for the development of CRC (Siegel, 2017).
Early detection and screening methods such as sigmoidoscopy, colonoscopy or stool-based tests for the detection of fecal blood (guaiac) or mutations (FIT-DNA) can reduce CRC deaths by detecting
cancers and removing polyps at early stages, but once the cancer develops, treatment is often surgical with chemotherapy. Newer drugs for targeted therapy include
EGFR inhibitors to slow the growth of the cancer, or
VEGF inhibitors to prevent the formation of blood vessels necessary for tumor growth.
IHC is used to identify CRC in tumors of unknown primary. CRCs express nuclear transcription factor CDX2,
which is highly specific for intestinal epithelial cells, and Villin, which is specific to adenocarcinomas of the GI tract.
GPA33 codes for a membranous protein that is expressed in a majority of colorectal tumors,
and it is an effective marker particularly for...
Click to Expand Text ▸
Primary IHC Markers In Colorectal Cancer
GPA33 (A33)
GPA33 (A33) gene codes for a membranous protein that is widely expressed in colorectal cancers, appearing in up to 95% of cases but particularly in well differentiated tumors. (Baptistella, 2016). Upregulation of GPA33 may be a downstream effect of peroxisome proliferator-activated receptor (PPARG) activation, which induces expression of KLF4, a transcription factor that targets GPA33 (Rageul, 2009). Antibodies to GPA33 used in the radiotherapy of colorectal cancers have been proposed as an effective means of treatment for tumors that express the protein (Cheal, 2017).
Staining:
Staining for this protein is typically membranous in well differentiated tumors and in normal tissue, but may be primarily cytoplasmic or nuclear in poorly differentiated and mucinous tumors (Baptistella, 2016).
LSBio's recommended antibody to GPA33 for use in immunohistochemistry is GPA33 Antibody LS‑B15164.
Click here for specifications and further information on this antibody.
Click here for specifications and further information on this antibody.
CK20 (KRT20)
CK20 (KRT20) is a keratin that is expressed in epithelial cells in colorectal crypts, where levels of this protein have been found to increase gradually from the crypt bottom (where it is absent) to the top (Chan, 2009; Moll, 1990). It is frequently used as a differentiation marker in the colon. Chan et al found that CK20 is positively regulated by CDX1, a homeobox gene involved in promoting intestinal differentiation (Chan, 2009). In colorectal cancer, CK20 typically has strong expression paired with an absence of expressed CK7 (KRT7) (Chan, 2009; Harbaum, 2012) and its expression is often inversely correlated with levels of LGR5 (Shimokawa, 2017). In a study on several hundred primary colorectal tumors, Harbaum et al found that while the majority had high levels of CK20, microsatellite unstable (MSI) tumors tended to have reduced or depleted expression of the protein (Harbaum, 2012). Aggressive, poorly differentiated colorectal tumors and those with high rates of MSI may stain negatively for this marker in immunohistochemistry (Harbaum, 2012; Merlos-Suárez, 2011).
Staining:
Staining is characteristically cytoplasmic.
LSBio's recommended antibody to CK20 for use in immunohistochemistry is CK20 Antibody LS‑B5959.
Click here for specifications and further information on this antibody.
Click here for specifications and further information on this antibody.
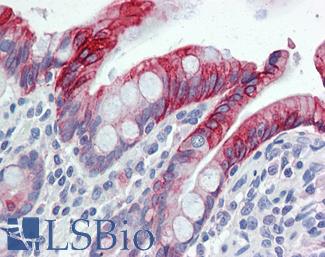
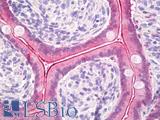
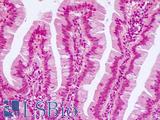
 Anti-KRT20 / Cytokeratin 20 antibody IHC of human small intestine. Immunohistochemistry of formalin-fixed, paraffin-embedded tissue after heat-induced antigen retrieval. Antibody LS-B5959 dilution 1:50. This image was taken for the unconjugated form of this product. Other forms have not been tested.
Anti-KRT20 / Cytokeratin 20 antibody IHC of human small intestine. Immunohistochemistry of formalin-fixed, paraffin-embedded tissue after heat-induced antigen retrieval. Antibody LS-B5959 dilution 1:50. This image was taken for the unconjugated form of this product. Other forms have not been tested.
CK20 Antibody LS-B5959
CK7 (KRT7)
CK7 (KRT7) is a keratin expressed in a number of tissues including in the colon, where its expression is limited to glandular cells (proteinatlas.org). CK7 typically has negative expression in colorectal cancer, and as a marker in IHC antibody panels it is often paired with positive expression of CK20 in support of adenocarcinoma (Harbaum, 2012; Diagnostic Immunohistochemistry p. 521). However, KRT7 expression in colon cancer may vary with the subtype and aggressiveness of the tumor; while high levels of CK20 expression are linked with increased differentiation, conversely in some colorectal tumors higher levels of CK7 and low levels of CK20 have been found and are attributed to poor differentiation (Harbaum, 2012; Harbaum, 2011). Furthermore, microsatellite-stable colorectal cancers with BRAF mutation, a subtype known to be aggressive and have poor prognosis, have been found to express higher levels of CK7 than other typically negative subtypes (Landau, 2014).
Staining:
Staining is characteristically cytoplasmic.
LSBio's recommended antibody to CK7 for use in immunohistochemistry is CK7 Antibody LS‑B7163.
Click here for specifications and further information on this antibody.
Click here for specifications and further information on this antibody.
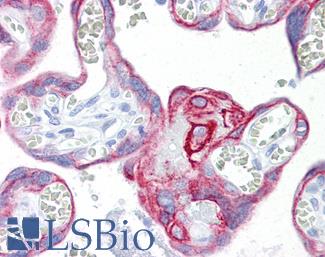
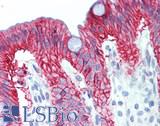
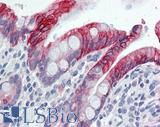
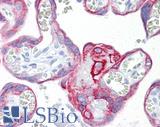 Anti-KRT7 / Cytokeratin 7 antibody IHC of human placenta. Immunohistochemistry of formalin-fixed, paraffin-embedded tissue after heat-induced antigen retrieval. Antibody LS-B7163 concentration 10 ug/ml.
Anti-KRT7 / Cytokeratin 7 antibody IHC of human placenta. Immunohistochemistry of formalin-fixed, paraffin-embedded tissue after heat-induced antigen retrieval. Antibody LS-B7163 concentration 10 ug/ml.
CK7 Antibody LS-B7163
Calretinin (CALB2)
Calretinin (CALB2) is a calcium-binding protein involved in calcium signaling. High levels of expression of calretinin in medullary carcinoma of the colon with strong focal staining in IHC have been documented, emphasizing its utility as an immunohistochemistry biomarker for detection of these cancers (Winn, 2008; Lin, 2014).
Staining:
Staining is can be cytoplasmic, but also potentially membranous and nuclear.
LSBio's recommended antibody to Calretinin for use in immunohistochemistry is Calretinin Antibody LS‑B4220.
Click here for specifications and further information on this antibody.
Click here for specifications and further information on this antibody.
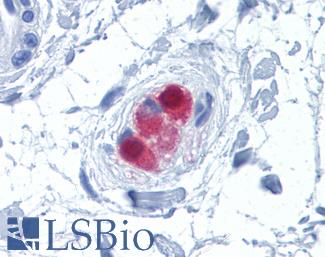
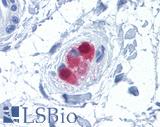 Anti-CALB2 / Calretinin antibody IHC of human colon, submucosal plexus. Immunohistochemistry of formalin-fixed, paraffin-embedded tissue after heat-induced antigen retrieval. Antibody LS-B4220 concentration 10 ug/ml.
Anti-CALB2 / Calretinin antibody IHC of human colon, submucosal plexus. Immunohistochemistry of formalin-fixed, paraffin-embedded tissue after heat-induced antigen retrieval. Antibody LS-B4220 concentration 10 ug/ml.
Calretinin Antibody LS-B4220
CDH17
CDH17 is a cadherin involved in cell adhesion and proliferation and expressed in the glands of the colon, duodenum and small intestine as well as the appendix and gallbladder (Fagerberg, 2014; proteinatlas.org). In a study examining over 700 tumors across multiple tissues including the colon, Panarelli et al found that CDH17 has immunohistochemistry staining patterns in gastrointestinal cancers similar to CDX2 and is a highly specific marker for the intestinal epithelium (Panarelli, 2012). Furthermore, overexpression of this protein has been implicated in metastasis and invasive potential in colorectal cancer (Bartolome, 2014).
Staining:
Staining is expected to be primarily membranous.
LSBio's recommended antibody to CDH17 for use in immunohistochemistry is CDH17 Antibody LS‑B6022.
Click here for specifications and further information on this antibody.
Click here for specifications and further information on this antibody.
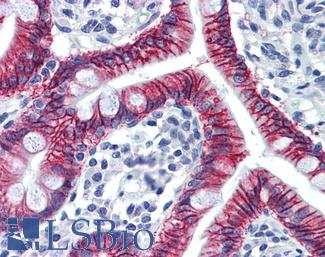
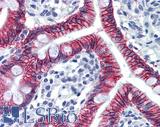 Anti-CDH17 / LI Cadherin antibody IHC of human small intestine. Immunohistochemistry of formalin-fixed, paraffin-embedded tissue after heat-induced antigen retrieval. Antibody LS-B6022 concentration 5 ug/ml.
Anti-CDH17 / LI Cadherin antibody IHC of human small intestine. Immunohistochemistry of formalin-fixed, paraffin-embedded tissue after heat-induced antigen retrieval. Antibody LS-B6022 concentration 5 ug/ml.
CDH17 Antibody LS-B6022
Cyclooxygenase 2 (COX-2 / PTGS2)
Cyclooxygenase 2 (COX-2 / PTGS2) is a protein involved in inflammation, proliferation and cell renewal that is regularly upregulated in colorectal cancer and may play a role in its progression (Liu, 2017; Wang, 2009). Colorectal cancer patients with tumors expressing high levels of COX-2 who are treated with NSAIDs and COXIBs (COX-2 inhibitors) see up to a 50% decrease in their risk of developing colorectal cancer (Wang, 2009). Mutant APC mouse models that undergo inhibition of this protein have also seen a decrease in tumorigenesis (Brown, 2005).
Staining:
Staining is expected to be cytoplasmic and potentially membranous.
LSBio's recommended antibody to COX-2 for use in immunohistochemistry is COX-2 Antibody LS‑B1608.
Click here for specifications and further information on this antibody.
Click here for specifications and further information on this antibody.
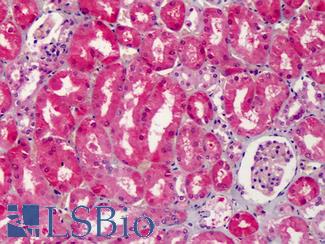
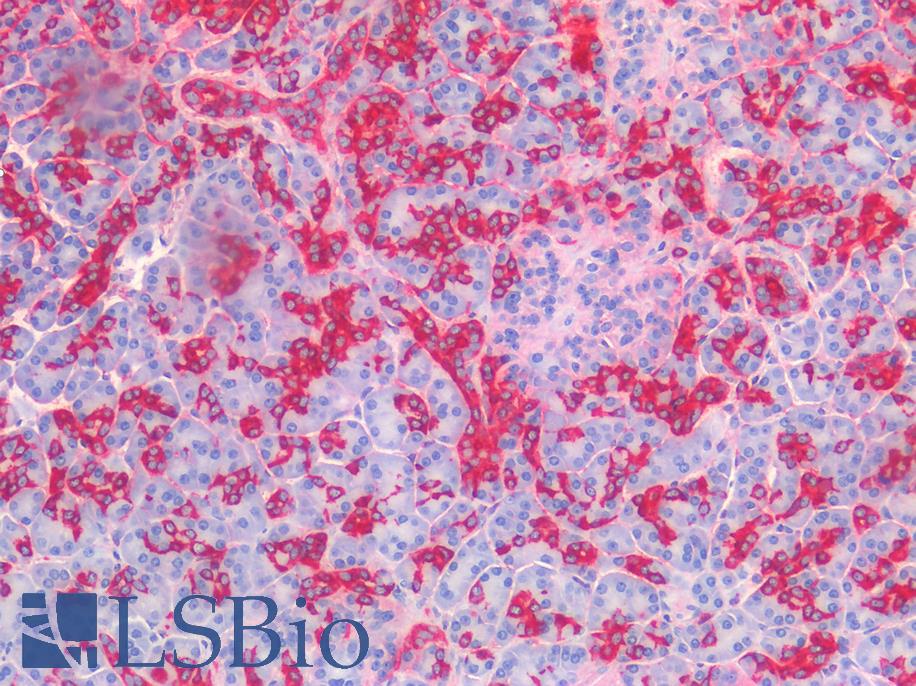
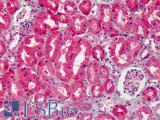 Anti-COX-2 antibody IHC of human kidney. Immunohistochemistry of formalin-fixed, paraffin-embedded tissue after heat-induced antigen retrieval. Antibody LS-B1608 concentration 5 ug/ml.
Anti-COX-2 antibody IHC of human kidney. Immunohistochemistry of formalin-fixed, paraffin-embedded tissue after heat-induced antigen retrieval. Antibody LS-B1608 concentration 5 ug/ml.
COX-2 Antibody LS-B1608
MUC2
MUC2 is a member of the mucin family of proteins and is a secreted protein that produces protective mucous in the gut (Allen, 1998). MUC2 is implicated in the progression of ulcerative colitis, where it is downregulated (Moehle, 2006), and also in the formation colorectal cancers (Betge, 2016). A MUC2 knockout model in mice demonstrated a propensity for colorectal cancer in its absence, as these mice frequently developed invasive colorectal adenocarcinomas (Velcich, 2002).
Staining:
Staining is expected to be cytoplasmic and potentially in secretions.
LSBio's recommended antibody to MUC2 for use in immunohistochemistry is MUC2 Antibody LS‑B5562.
Click here for specifications and further information on this antibody.
Click here for specifications and further information on this antibody.
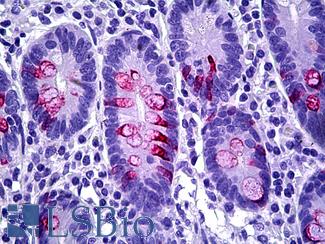
 Anti-MUC2 antibody IHC of human intestine. Immunohistochemistry of formalin-fixed, paraffin-embedded tissue after heat-induced antigen retrieval. Antibody LS-B5562 dilution 1:100.
Anti-MUC2 antibody IHC of human intestine. Immunohistochemistry of formalin-fixed, paraffin-embedded tissue after heat-induced antigen retrieval. Antibody LS-B5562 dilution 1:100.
MUC2 Antibody LS-B5562
Cell-Type Specific Markers In Colorectal Cancer
CDX2
CDX2 codes for a homeobox protein involved in intestinal cell differentiation and tumor suppression in the colon (Platet, 2017), and loss of expression of this protein is correlated with colorectal tumorigenesis and poor survival (Platet, 2017; Dalerba, 2016). Platet et al found that higher levels of expression in the colon are correlated with a lack of cellular deformability and an increase in anti-metastatic structural properties (Platet, 2017). Furthermore, loss of expression of this protein may lead to tumor cells attaining stem cell attributes, as described by Lundberg IV et al in their findings that CDX2 downregulation is linked with upregulation of Yamanaka factor SOX2, involved in embryonic development and implicated in the induction of pluripotency (Lundberg, 2016; Takahashi, 2006). They found this to be a frequent phenomenon in MSI-positive (microsatellite unstable) and CIMP (CpG Island Methylator Phenotype) tumors. Jiang et al also found a link between methylation silencing of CDX2 (hypermethylation is a driving factor of CIMP) and colorectal cancer (Jiang, 2016). Downregulation of CDX2 may also be caused by FBXW7, a tumor suppressor with frequent mutations in cancer that manages the degradation of a large number of proteins including the Yamanaka factor Myc (Kumar, 2016; Takeishi, 2014). FBXW7 has been found to negatively regulate CDX2 expression by promoting its degradation together with the serine-threonine kinase GSK3B (Kumar, 2016).
Staining:
Staining is expected to be nuclear.
LSBio's recommended antibody to CDX2 for use in immunohistochemistry is CDX2 Antibody LS‑B1514.
Click here for specifications and further information on this antibody.
Click here for specifications and further information on this antibody.
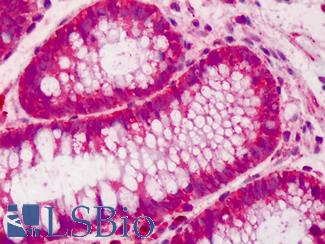
 Anti-CDX2 antibody IHC of human colon. Immunohistochemistry of formalin-fixed, paraffin-embedded tissue after heat-induced antigen retrieval. Antibody LS-B1514 concentration 5 ug/ml. This image was taken for the unconjugated form of this product. Other forms have not been tested.
Anti-CDX2 antibody IHC of human colon. Immunohistochemistry of formalin-fixed, paraffin-embedded tissue after heat-induced antigen retrieval. Antibody LS-B1514 concentration 5 ug/ml. This image was taken for the unconjugated form of this product. Other forms have not been tested.
CDX2 Antibody LS-B1514
Villin
Villin (VIL1) is an actin-binding protein involved in the maintenance of microvilli in epithelial cells and is also involved in the epithelial-mesenchymal transition, and deregulation of this gene may contribute to tumorigenesis (Patnaik, 2016). Loss of expression of villin is seen in poorly differentiated colorectal carcinomas, particularly in microsatellite-unstable (MSI) tumors (Arango, 2012), and is linked to poor survival (Jaudah, 2013).
Staining:
Staining is expected to localize to luminal membranes.
LSBio's recommended antibody to Villin for use in immunohistochemistry is Villin Antibody LS‑B8547.
Click here for specifications and further information on this antibody.
Click here for specifications and further information on this antibody.
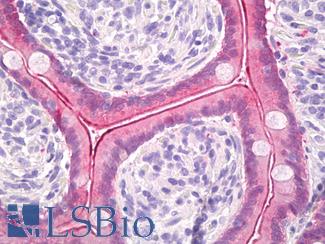
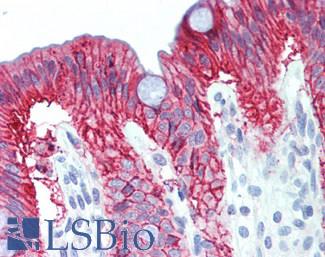
 Anti-Villin antibody IHC of human small intestine, epithelial membrane. Immunohistochemistry of formalin-fixed, paraffin-embedded tissue after heat-induced antigen retrieval. Antibody LS-B8547 dilution 10 ug/ml.
Anti-Villin antibody IHC of human small intestine, epithelial membrane. Immunohistochemistry of formalin-fixed, paraffin-embedded tissue after heat-induced antigen retrieval. Antibody LS-B8547 dilution 10 ug/ml.
Villin Antibody LS-B8547
MOC-31 (EPCAM)
MOC-31 is a clone of the protein EPCAM (TACSTD1) and is a useful biotarget for metastatic adenocarcinomas, including those originating in the colon. As an immunomarker, MOC-31 antibodies are used to distinguish adenocarcinoma from mesothelial cells with high sensitivity (Morgan, 1999; Kundu, 2011; Hecht, 2006). EPCAM itself is a protein involved in cell adhesion and has been implicated in the progression of a number of cancers (Dai, 2017; Baeuerle, 2007), included a proportion of individuals with Lynch Syndrome who hold inherited deletions in the gene. These deletions lead to silencing of the adjacent repair gene MSH2 via transcriptional read-through, which then causes microsatellite instability and colorectal cancer (as well as other malignancies that also frequently arise from Lynch Syndrome) (Kempers, 2010).
Staining:
Staining is expected to be primarily membranous.
LSBio's recommended antibody to EPCAM / MOC‑31 for use in immunohistochemistry is EPCAM / MOC‑31 Antibody LS‑B5565.
Click here for specifications and further information on this antibody.
Click here for specifications and further information on this antibody.
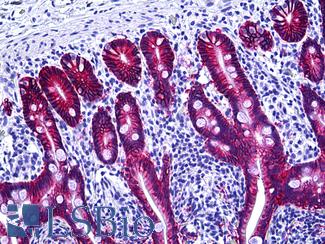
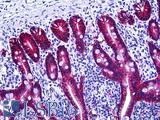 Anti-EPCAM antibody IHC of human intestine. Immunohistochemistry of formalin-fixed, paraffin-embedded tissue after heat-induced antigen retrieval. Antibody LS-B5565 dilution 1:100.
Anti-EPCAM antibody IHC of human intestine. Immunohistochemistry of formalin-fixed, paraffin-embedded tissue after heat-induced antigen retrieval. Antibody LS-B5565 dilution 1:100.
EPCAM / MOC‑31 Antibody LS-B5565
Metastatic Markers In Colorectal Cancer
CEA / Carcinoembryonic Antigen
Carcinoembryonic (CEA / mCEA / pCEA) antigen includes a large class of proteins such as CEACAM5 that are biomarkers for colorectal cancer. These cell adhesion proteins have high expression in specifically metastatic cancer, making them suitable targets for immunohistochemistry of late tumors (Bajenova, 2014; Polat, 2014). The degree of expression of CEA is correlated with tumor stage, with particularly pronounced levels in well-differentiated adenocarcinomas relative to early tumors (which are expected to express the same levels as normal tissues) (Vukobrat-Bijedic, 2013).
Staining:
Staining is expected to be membranous and cytoplasmic.
LSBio's recommended antibody to CEA for use in immunohistochemistry is CEA Antibody LS‑B7173.
Click here for specifications and further information on this antibody.
Click here for specifications and further information on this antibody.
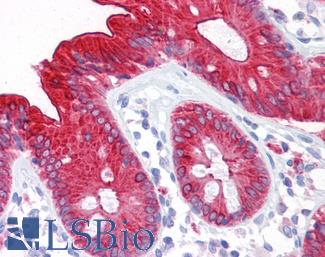
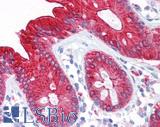 Anti-CEA antibody IHC of human colon. Immunohistochemistry of formalin-fixed, paraffin-embedded tissue after heat-induced antigen retrieval. Antibody LS-B7173 concentration 10 ug/ml.
Anti-CEA antibody IHC of human colon. Immunohistochemistry of formalin-fixed, paraffin-embedded tissue after heat-induced antigen retrieval. Antibody LS-B7173 concentration 10 ug/ml.
CEA Antibody LS-B7173
SATB2
SATB2 is a transcription factor involved in chromatin remodeling that negatively regulates a number of stem cell markers such as AXIN2, CD44 and NANOG (Li, 2016; Dragomir, 2014). Loss of expression of SATB2 in colorectal cancer has been correlated with an increase in stemness and invasive potential (1). SATB2 may be a useful immunohistochemistry (IHC) marker for primary and metastatic colorectal cancers due to its high specificity (Dragomir, 2014).
Staining:
Staining is expected to be primarily nuclear.
LSBio's recommended antibody to SATB2 for use in immunohistochemistry is SATB2 Antibody LS‑B4981.
Click here for specifications and further information on this antibody.
Click here for specifications and further information on this antibody.
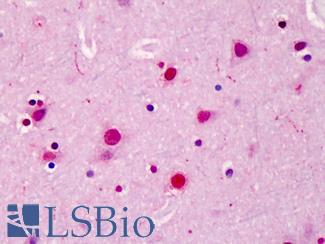
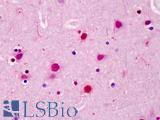 Anti-SATB2 antibody IHC of human brain, cortex. Immunohistochemistry of formalin-fixed, paraffin-embedded tissue after heat-induced antigen retrieval. Antibody LS-B4981 concentration 5 ug/ml.
Anti-SATB2 antibody IHC of human brain, cortex. Immunohistochemistry of formalin-fixed, paraffin-embedded tissue after heat-induced antigen retrieval. Antibody LS-B4981 concentration 5 ug/ml.
SATB2 Antibody LS-B4981
Carbohydrate antigen (CA19-9/FUT)
Carbohydrate antigen (CA19-9/FUT) is a common metastatic colorectal cancer marker to tetrasaccharide carbohydrate (Sialyl-Lewis A), which has high levels of occurrence alongside CEACAM5 (CEA) expression levels in late tumors (Vukobrat-Bijedic, 2013; Polat, 2014). CA19-9 is an effective marker for circulating tumor cells, as increased levels of CA19-9 in serum coincide with the quantity of circulating tumor cells from colorectal metastases (Zhao, 2017).
Staining:
Staining is primarily cytoplasmic.
LSBio's recommended antibody to CA19-9 for use in immunohistochemistry is CA19-9 Antibody LS‑B5366.
Click here for specifications and further information on this antibody.
Click here for specifications and further information on this antibody.
Oncogenic Markers (Wild Type) In Colorectal Cancer
TP53
TP53 is a tumor suppressor, repair gene and transcriptional regulator, and it is considered the most frequently mutated gene in cancer (Andrysik, 2017). Germline mutations in TP53 lead to Li-Fraumeni syndrome and cancer, including early-onset colorectal cancer (Yurgelun, 2015). In colorectal cancer, the expression of TP53 with gain of function mutations is a regular occurrence and is correlated with an increase in proliferative and invasive attributes in tumors of advanced stage (Andrysik, 2017; Sun, 2016). Furthermore, TP53 is often overexpressed in colorectal dysplasia resulting from ulcerative colitis (Kobayashi, 2017).
Staining:
Staining is expected to be nuclear.
LSBio's recommended antibody to TP53 for use in immunohistochemistry is TP53 Antibody LS‑B7722.
Click here for specifications and further information on this antibody.
Click here for specifications and further information on this antibody.
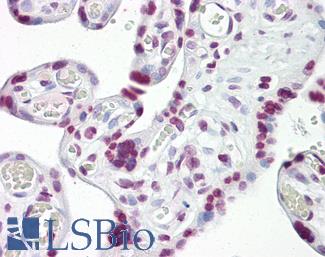
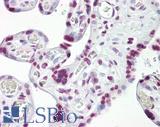 Anti-p53 antibody IHC of human placenta. Immunohistochemistry of formalin-fixed, paraffin-embedded tissue after heat-induced antigen retrieval. Antibody LS-B7722 dilution 10 ug/ml.
Anti-p53 antibody IHC of human placenta. Immunohistochemistry of formalin-fixed, paraffin-embedded tissue after heat-induced antigen retrieval. Antibody LS-B7722 dilution 10 ug/ml.
TP53 Antibody LS-B7722
BRAF
BRAF, a MAPK/ERK signaling pathway kinase, is one of the most frequently somatically mutated genes in some cancers, particularly melanoma, thyroid carcinomas, and colorectal cancer (Chang, 2016; Zhang, 2014; Cantwell-Dorris, 2011). BRAF mutation occurs in roughly 10% of colorectal cancers, where detection of the V600E (rs113488022) mutation is useful for subtyping CIMP (CpG Island Methylator Phenotype), particularly when it accompanies the disappearance of the repair protein MLH1 (Cantwell-Dorris, 2011). These microsatellite-unstable (MSI) tumors typically propagate along the sessile serrated adenoma development pathway (Rhee, 2016; Cantwell-Dorris, 2011) and experience epigenetic silencing and loss of function frameshift in a large number of tumor suppressor and cell cycle regulation genes including TGFBR2, BAX, RNF43, CASP5 and KMT2C (MLL3) (Cortes-Ciriano, 2017). This is caused by the hypermethylation-induced silencing of mismatch repair protein MLH1, which occurs subsequent to and may even be caused by BRAF mutation (Fang M, 2014; Cantwell-Dorris, 2011). BRAF V600E is a pro-proliferative mutation, and while the adoption of V600E-mutant BRAF protein alone is not a pathogenic event (this mutation is the source of overproliferation in many benign nevi) (Wu, 2007), when combined with mutations in tumor suppressor genes it leads to carcinogenesis.
Staining:
Staining may be nuclear, cytoplasmic or membranous.
LSBio's recommended antibody to BRAF for use in immunohistochemistry is BRAF Antibody LS‑B1627.
Click here for specifications and further information on this antibody.
Click here for specifications and further information on this antibody.
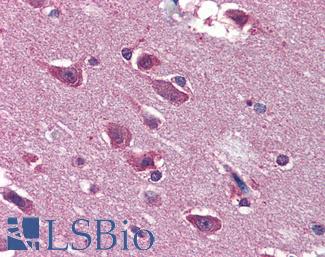
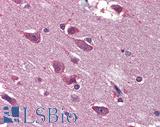 Anti-BRAF antibody IHC of human brain, cortex. Immunohistochemistry of formalin-fixed, paraffin-embedded tissue after heat-induced antigen retrieval. Antibody LS-B3862 dilution 1:200.
Anti-BRAF antibody IHC of human brain, cortex. Immunohistochemistry of formalin-fixed, paraffin-embedded tissue after heat-induced antigen retrieval. Antibody LS-B3862 dilution 1:200.
BRAF Antibody LS-B1627
PIK3CA
PIK3CA is a kinase frequently mutated in cancer, including up to 15% of colorectal cancers. This gene is regularly mutated in proximal cancers alongside KRAS mutation and MGMT silencing (Rosty, 2013), and frequently experiences the same somatic mutations characterized by H1047R, E545K, and E542K (Chang, 2016). The acquisition of somatic mutations in this gene may lower rates of survival in drug-resistant colorectal tumors that also harbor KRAS-mutations (Xu, 2017).
Staining:
Staining is expected to be primarily cytoplasmic but ay also localize to membranes.
LSBio's recommended antibody to PIK3CA for use in immunohistochemistry is PIK3CA Antibody LS‑B5363.
Click here for specifications and further information on this antibody.
Click here for specifications and further information on this antibody.
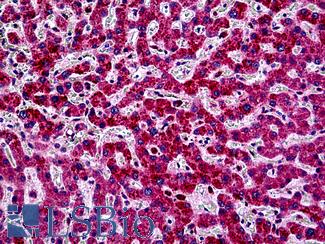
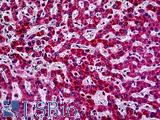 Anti-PIK3CA antibody IHC of human liver. Immunohistochemistry of formalin-fixed, paraffin-embedded tissue after heat-induced antigen retrieval. Antibody LS-B5363 concentration 5 ug/ml.
Anti-PIK3CA antibody IHC of human liver. Immunohistochemistry of formalin-fixed, paraffin-embedded tissue after heat-induced antigen retrieval. Antibody LS-B5363 concentration 5 ug/ml.
PIK3CA Antibody LS-B5363
RECQL4
RECQL4 is a RECQ-like helicase involved in aging and DNA repair that forms a complex with TP53 (Qiao, 2016; Croteau, 2012). This protein has mutations, amplified copy numbers and upregulation in a number of cancers including colorectal, breast and gastric cancer (Qiao, 2016; Cancer Genome Atlas Network, 2012; Mo, 2016; Arora, 2016; Lao, 2013; Buffart, 2005; Giannakis, 2016). Amplification of this protein in metastatic colorectal cancer seems to be correlated with other oncogenes on chromosome 8, including Yamanaka factor MYC, SLA, PTK2, PTP4A3, TPD52, MOS (Buffart, 2005), as well as PLEC, EPPK1, RAD21, and PDS5B (Cancer Genome Atlas Network, 2012; Buffart, 2005; Giannakis, 2016). In gastric cancer cells, high levels of expression of RECQL4 have been linked to drug resistance and activation of the multi-drug resistant protein 1 (MDR1 / P-glycoprotein 1 / ABCB1 / CD243) (Mo, 2016). IHC (immunohistochemistry) staining of RECQL4 in colorectal cancer showed this protein localizing to the nucleus (Lao, 2013).
Staining:
Staining is expected to be predominantly nuclear.
LSBio's recommended antibody to RECQL4 for use in immunohistochemistry is RECQL4 Antibody LS‑B13621.
Click here for specifications and further information on this antibody.
Click here for specifications and further information on this antibody.
Lynch and FAP Related Markers
Adenomatous Polyposis Coli (APC)
Adenomatous Polyposis Coli (APC) is a gene frequently mutated in colorectal cancer and inherited mutations in this gene are causative for classical familial adenomatous polyposis. This gene is involved in the regulation of cell adhesion through its interaction with CDH1 (e-cadherin) and CTNNB1 (beta-catenin) via the WNT signaling pathway (Markowitz, 2009), and it is considered a tumor suppressor. The loss of functional APC protein (often alongside mutations in TP53 or KRAS) is a frequent event in colorectal cancers and is associated with cellular over-proliferation, chromosomal instability and carcinogenesis (Pino, 2010).
Staining:
Staining is expected to be primarily cytoplasmic.
LSBio's recommended antibody to APC for use in immunohistochemistry is APC Antibody LS‑B2788.
Click here for specifications and further information on this antibody.
Click here for specifications and further information on this antibody.
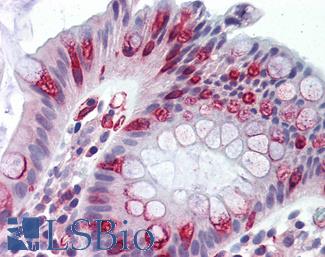
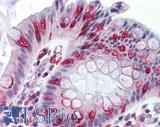 Anti-APC antibody IHC of human colon. Immunohistochemistry of formalin-fixed, paraffin-embedded tissue after heat-induced antigen retrieval. Antibody LS-B2788 concentration 10 ug/ml.
Anti-APC antibody IHC of human colon. Immunohistochemistry of formalin-fixed, paraffin-embedded tissue after heat-induced antigen retrieval. Antibody LS-B2788 concentration 10 ug/ml.
APC Antibody LS-B2788
MLH1
MLH1 is a primary MMR (mismatch repair) DNA repair gene, and loss of function of MLH1 protein leads to microsatellite instability and cancer in the colon and other organs (Papadopoulos, 1994). This can be caused by a “second hit” somatic mutation in MLH1 in patients with inherited heterozygous pathogenic mutations (Ollikainen, 2007), or alternatively through epigenetic silencing via hypermethylation of the MLH1 promoter in CIMP (CpG Island Methylator Phenotype) colorectal tumors (Boland, 2010). Inherited MLH1 mutation is one of the most frequent causes of Lynch Syndrome, and it is a pivotal gene in the mismatch repair functionality of cells (Cohen, 2014). IHC staining for MLH1 is therefore useful for determining the tumor subtype of colorectal cancer patients, as in Lynch Syndrome, Lynch-like cancers and in CIMP cases expression of this protein regularly disappears.
Staining:
Staining is expected to be nuclear.
LSBio's recommended antibody to MLH1 for use in immunohistochemistry is MLH1 Antibody LS‑B3475.
Click here for specifications and further information on this antibody.
Click here for specifications and further information on this antibody.
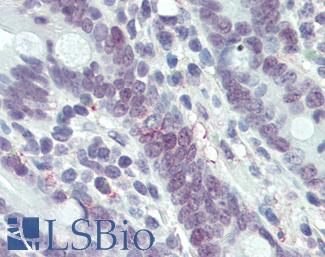
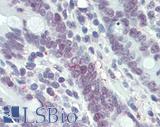 Anti-MLH1 antibody IHC of human small intestine. Immunohistochemistry of formalin-fixed, paraffin-embedded tissue after heat-induced antigen retrieval. Antibody LS-B3475 dilution 1:200.
Anti-MLH1 antibody IHC of human small intestine. Immunohistochemistry of formalin-fixed, paraffin-embedded tissue after heat-induced antigen retrieval. Antibody LS-B3475 dilution 1:200.
MLH1 Antibody LS-B3475
MSH2
MSH2 is a DNA repair protein involved in mismatch repair and inherited or somatic mutations in this gene are a source of microsatellite instability (MSI) and cancer in the colon. Heterozygous germline mutations in MSH2 are the cause of disease in a large subset of Lynch Syndrome (LS) patients (Cohen, 2014); in LS, somatic mutation of the remaining functional copy of this protein leads to MSI, widespread mutation and LOF in a number of important tumor suppressor and apoptotic genes, and subsequently tumorigenesis throughout the colon (Lynch, 2009).
Staining:
Staining is expected to be nuclear.
LSBio's recommended antibody to MSH2 for use in immunohistochemistry is MSH2 Antibody LS‑B1973.
Click here for specifications and further information on this antibody.
Click here for specifications and further information on this antibody.
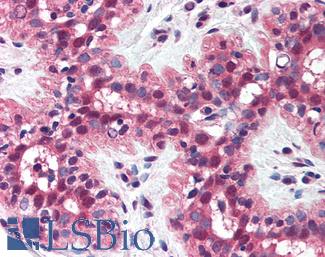
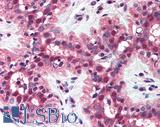 Anti-MSH2 antibody IHC of human breast. Immunohistochemistry of formalin-fixed, paraffin-embedded tissue after heat-induced antigen retrieval. Antibody LS-B1973 concentration 5 ug/ml.
Anti-MSH2 antibody IHC of human breast. Immunohistochemistry of formalin-fixed, paraffin-embedded tissue after heat-induced antigen retrieval. Antibody LS-B1973 concentration 5 ug/ml.
MSH2 Antibody LS-B1973
MSH6
MSH6 is a DNA repair protein involved specifically in the mismatch repair of mutations that arise from replication and recombination, and somatic and inherited mutations in this gene lead to microsatellite instability and colorectal cancer. MSH6, along with MSH2, MSH3, MLH1, PMS1 and PMS2, is dysfunctional in the germline of many Lynch Syndrome patients (Houlleberghs, 2017; Giglia, 2016), and genetic testing along with immunohistochemistry staining of this protein is useful for diagnosis and subtyping of colorectal tumors (Giglia, 2016).
Staining:
Staining is expected to be nuclear.
LSBio's recommended antibody to MSH6 for use in immunohistochemistry is MSH6 Antibody LS‑B10739.
Click here for specifications and further information on this antibody.
Click here for specifications and further information on this antibody.
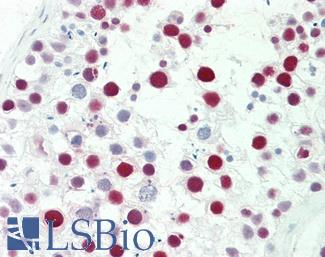
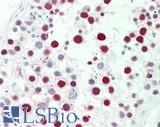 Anti-MSH6 antibody IHC staining of human testis. Immunohistochemistry of formalin-fixed, paraffin-embedded tissue after heat-induced antigen retrieval. Antibody LS-B10739 dilution 1:50.
Anti-MSH6 antibody IHC staining of human testis. Immunohistochemistry of formalin-fixed, paraffin-embedded tissue after heat-induced antigen retrieval. Antibody LS-B10739 dilution 1:50.
MSH6 Antibody LS-B10739
PMS2
PMS2 is a DNA mismatch repair protein that is often mutated or downregulated in colorectal cancer. Inherited mutations in PMS2 are the cause of Lynch Syndrome and colorectal cancer in a subset of patients (Rosty, 2016; Cohen, 2014). Knockout of PMS2 results in microsatellite unstable tumors with malignant potential, and IHC staining for this protein can thus be used to subtype colorectal tumors (Worthley, 2005).
Staining:
Staining is expected to be nuclear.
LSBio's recommended antibody to PMS2 for use in immunohistochemistry is PMS2 Antibody LS‑B1980.
Click here for specifications and further information on this antibody.
Click here for specifications and further information on this antibody.
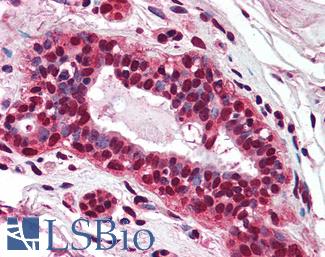
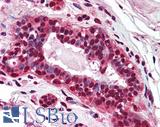 Anti-PMS2 antibody IHC of human breast. Immunohistochemistry of formalin-fixed, paraffin-embedded tissue after heat-induced antigen retrieval. Antibody LS-B1980 concentration 5 ug/ml.
Anti-PMS2 antibody IHC of human breast. Immunohistochemistry of formalin-fixed, paraffin-embedded tissue after heat-induced antigen retrieval. Antibody LS-B1980 concentration 5 ug/ml.
PMS2 Antibody LS-B1980
WNT-Signaling Pathway Markers
AXIN1
AXIN1 is a protein involved in the WNT signaling pathway and acts alongside APC, CK1, AXIN2 and GSK3B in the regulation of B-Catenin (CTNNB1) phosphorylation and degradation (Mazzoni, 2014; Novellasdemunt, 2015). Mutations in this gene are prevalent in microsatellite-unstable (MSI) colorectal cancers, where loss of function of this protein may have a negative activating impact on WNT signaling and B-Catenin management and contribute to tumorigenesis (Novellasdemunt, 2015; Segditsas, 2006).
Staining:
Staining may be cytoplasmic, nuclear and/or membranous.
LSBio's recommended antibody to AXIN1 for use in immunohistochemistry is AXIN1 Antibody LS‑B2741.
Click here for specifications and further information on this antibody.
Click here for specifications and further information on this antibody.
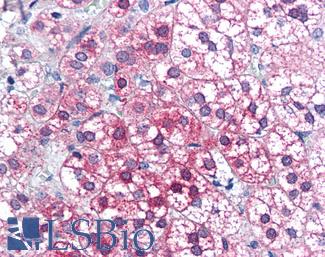
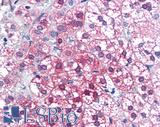 Anti-AXIN1 / AXIN antibody IHC of human adrenal. Immunohistochemistry of formalin-fixed, paraffin-embedded tissue after heat-induced antigen retrieval. Antibody LS-B2741 concentration 75 ug/ml.
Anti-AXIN1 / AXIN antibody IHC of human adrenal. Immunohistochemistry of formalin-fixed, paraffin-embedded tissue after heat-induced antigen retrieval. Antibody LS-B2741 concentration 75 ug/ml.
AXIN1 Antibody LS-B2741
AXIN2
AXIN2, also known as conductin, is a member of the WNT signaling pathway and maintains the degradation and phosphorylation of B-Catenin (CTNNB1) alongside APC, AXIN1, CK1 and CSK3B in basal colorectal cells (Mazzoni, 2014; Novellasdemunt, 2015; Segditsas, 2006). AXIN2 is somatically mutated in microsatellite-unstable (MSI) colorectal cancers, where loss of function contributes to aberrant activation of WNT signaling and tumorigenesis. Germline mutations in this gene are also associated with a predisposition for colorectal carcinoma (Segditsas, 2006).
Staining:
Staining may be cytoplasmic, nuclear and/or membranous.
LSBio's recommended antibody to AXIN2 for use in immunohistochemistry is AXIN2 Antibody LS‑B7029.
Click here for specifications and further information on this antibody.
Click here for specifications and further information on this antibody.
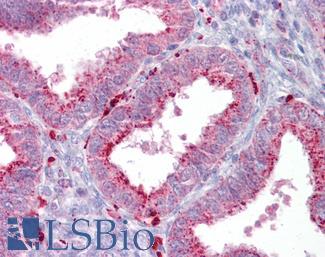
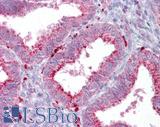 Anti-AXIN2 / AXIL antibody IHC of human uterus. Immunohistochemistry of formalin-fixed, paraffin-embedded tissue after heat-induced antigen retrieval. Antibody LS-B7029 concentration 5 ug/ml.
Anti-AXIN2 / AXIL antibody IHC of human uterus. Immunohistochemistry of formalin-fixed, paraffin-embedded tissue after heat-induced antigen retrieval. Antibody LS-B7029 concentration 5 ug/ml.
AXIN2 Antibody LS-B7029
TCF7L2
TCF7L2 is a member of the WNT signaling pathway and interacts with B-catenin. It is frequently mutated in MSI- (microsatellite instability) positive colorectal tumors, and also forms fusion transcripts with the neighboring VTI1A gene in some colorectal cancers (Nome, 2014). Homozygous germline mutations in this gene (mutations rs12255372 and rs7903146) have been associated with a risk for colorectal cancer as well (Folsom, 2008; Sainz, 2012; Hazra, 2008).
Staining:
Staining may be nuclear and cytoplasmic.
LSBio's recommended antibody to TCF7L2 for use in immunohistochemistry is TCF7L2 Antibody LS‑B7129.
Click here for specifications and further information on this antibody.
Click here for specifications and further information on this antibody.
References ▷
-
Allen, Adrian et al. The MUC2 Gene Product: A Human Intestinal Mucin. The International Journal of Biochemistry & Cell Biology 30 (7):797–801. 7/1/1998
[Full Text Article] -
Al-Maghrabi, Jaudah et al. Loss of Villin Immunoexpression in Colorectal Carcinoma Is Associated with Poor Differentiation and Survival. ISRN Gastroenterology 2013:7. 2013
-
Arango, Diego et al. Villin Expression Is Frequently Lost in Poorly Differentiated Colon Cancer. The American Journal of Pathology 180 (4):1509–21. 2012/04
[Full Text Article] [Pubmed: 22349300] [PMC: PMC5426554] -
Arora, Arvind et al. RECQL4 Helicase Has Oncogenic Potential in Sporadic Breast Cancers. The Journal of Pathology 238 (4):495–501. 2016/03
[Full Text Article] [Pubmed: 26690729] [PMC: PMC5576893] -
Baeuerle, P. A. et al. EpCAM (CD326) Finding Its Role in Cancer. British Journal of Cancer 96 (3):417–23. 2/12/2007
[Full Text Article] [Pubmed: 17211480] [PMC: PMC2360029] -
Bajenova, Olga et al. Carcinoembryonic Antigen Promotes Colorectal Cancer Progression by Targeting Adherens Junction Complexes. Experimental Cell Research 324 (2):115–23. 6/10/2014
[Full Text Article] -
Baptistella, Antuani R. et al. Heterogeneous Expression of A33 in Colorectal Cancer: Possible Explanation for A33 Antibody Treatment Failure. Anti-Cancer Drugs 27 (8):734–37. 2016
[Full Text Article] [Pubmed: 27272411] -
Bartolome, R. A. et al. Cadherin-17 Interacts with Alpha2beta1 Integrin to Regulate Cell Proliferation and Adhesion in Colorectal Cancer Cells Causing Liver Metastasis. Oncogene 33 (13):1658–69. 3/27/2014
[Full Text Article] [Pubmed: 23604127] -
Boland, C. Richard et al. Microsatellite Instability in Colorectal Cancer. Gastroenterology 138 (6):2073–2087.e3. 2010/06
[Full Text Article] [Pubmed: 20420947] [PMC: PMC3037515] -
Brown, Joanne R. et al. COX-2: A Molecular Target for Colorectal Cancer Prevention. Journal of Clinical Oncology 23 (12):2840–55. 4/20/2005
[Full Text Article] -
Buffart, T. E. et al. DNA Copy Number Changes at 8q11-24 in Metastasized Colorectal Cancer. Cellular Oncology : The Official Journal of the International Society for Cellular Oncology 27 (1):57–65. 2005
[Pubmed: 15750208] [PMC: PMC4611113] -
Cantwell-Dorris, Emma R. et al. BRAF V600E: Implications for Carcinogenesis and Molecular Therapy. Molecular Cancer Therapeutics 10 (3):385. 3/1/2011
[Full Text Article] -
Chang, Matthew T. et al. Identifying Recurrent Mutations in Cancer Reveals Widespread Lineage Diversity and Mutational Specificity. Nature Biotechnology 34 (2):155–63. 2016/02
[Full Text Article] [Pubmed: 26619011] [PMC: PMC4744099] -
Cheal, Sarah M. et al. Curative Multicycle Radioimmunotherapy Monitored by Quantitative SPECT/CT-Based Theranostics, Using Bispecific Antibody Pretargeting Strategy in Colorectal Cancer. Journal of Nuclear Medicine 58 (11):1735–42. 11/1/2017
[Full Text Article] [Pubmed: 28705917] -
Chen, Wang Yang et al. Chromobox Homolog 2 Protein: A Novel Biomarker for Predicting Prognosis and Taxol Sensitivity in Patients with Breast Cancer. Oncology Letters 13 (3):1149–56. 2017/03
[Full Text Article] [Pubmed: 28454227] [PMC: PMC5403172] -
Choi, Yong Won et al. B-RafV600E Inhibits Sodium Iodide Symporter Expression via Regulation of DNA Methyltransferase 1. Experimental & Molecular Medicine 46 (11):e120. 2014/11
[Full Text Article] -
Cortes-Ciriano, Isidro et al. A Molecular Portrait of Microsatellite Instability across Multiple Cancers. Nature Communications 8 (June):15180. 6/6/2017
[Full Text Article] -
Dalerba, Piero et al. CDX2 as a Prognostic Biomarker in Stage II and Stage III Colon Cancer. The New England Journal of Medicine 374 (3):211–22. 1/21/2016
[Full Text Article] [Pubmed: 26789870] [PMC: PMC4784450] -
Dragomir, Anca et al. The Role of SATB2 as a Diagnostic Marker for Tumors of Colorectal OriginResults of a Pathology-Based Clinical Prospective Study. American Journal of Clinical Pathology 141 (5):630–38. 5/1/2014
[Full Text Article] -
Fagerberg, Linn et al. Analysis of the Human Tissue-Specific Expression by Genome-Wide Integration of Transcriptomics and Antibody-Based Proteomics. Molecular & Cellular Proteomics : MCP 13 (2):397–406. 2014/02
[Full Text Article] [Pubmed: 24309898] [PMC: PMC3916642] -
Giannakis, Marios et al. Genomic Correlates of Immune-Cell Infiltrates in Colorectal Carcinoma. Cell Reports, April. 4/14/2016
[Full Text Article] [Pubmed: 27149842] [PMC: PMC4850357] -
Guo, Jianhui et al. Mutations in the Human Naked Cuticle Homolog NKD1 Found in Colorectal Cancer Alter Wnt/Dvl/Beta-Catenin Signaling. PloS One 4 (11):e7982. 11/24/2009
[Full Text Article] [Pubmed: 19956716] [PMC: PMC2776356] -
Harbaum, Lars et al. Keratin 20 - a Diagnostic and Prognostic Marker in Colorectal Cancer? Histology and Histopathology 27 (3):347–56. 2012/03
[Full Text Article] [Pubmed: 22237712] -
Harbaum, Lars et al. Keratin 7 Expression in Colorectal Cancer--Freak of Nature or Significant Finding? Histopathology 59 (2):225–34. 2011/08
[Full Text Article] [Pubmed: 21884201] -
Hause, Ronald J. et al. Classification and Characterization of Microsatellite Instability across 18 Cancer Types. Nature Medicine 22 (11):1342–50. 2016
[Full Text Article] [Pubmed: 27694933] -
Hecht, Jonathan L. et al. Monoclonal Antibody MOC-31 Reactivity as a Marker for Adenocarcinoma in Cytologic Preparations. Cancer 108 (1):56–59. 2/25/2006
[Full Text Article] [Pubmed: 16329115] -
Huang, Lei et al. Copy Number Alterations in Colorectal Cancer. International Journal of Clinical and Experimental Medicine 10 (1):204–14. 2017
-
Jansen, Anne ML et al. Combined Mismatch Repair and POLE/POLD1 Defects Explain Unresolved Suspected Lynch Syndrome Cancers. European Journal of Human Genetics 24 (7):1089–92. 2016/07
[Full Text Article] [Pubmed: 26648449] [PMC: PMC5070903] -
Jiang, Guozhong et al. Methylation of CDX2 as a Predictor in Poor Clinical Outcome of Patients with Colorectal Cancer. Genetic Testing and Molecular Biomarkers 20 (11):710–14. 2016/11
[Full Text Article] [Pubmed: 27754705] -
Kandalaft, Patricia L. et al. Practical Applications in Immunohistochemistry: Carcinomas of Unknown Primary Site. Archives of Pathology & Laboratory Medicine 140 (6):508–23. 2016/06
[Full Text Article] [Pubmed: 26457625] -
Kumar, Yogesh et al. Ubiquitin Ligase, Fbw7, Targets CDX2 for Degradation via Two Phosphodegron Motifs in a GSK3beta-Dependent Manner. Molecular Cancer Research : MCR 14 (11):1097–1109. 2016/11
[Full Text Article] [Pubmed: 27470268] -
Kundu, Uma R. et al. Use of the Monoclonal Antibody MOC-31 as an Immunomarker for Detecting Metastatic Adenocarcinoma in Effusion Cytology. Cancer Cytopathology 119 (4):272–78. 8/25/2011
[Full Text Article] [Pubmed: 21732548] -
Landau, Michael S. et al. BRAF-Mutated Microsatellite Stable Colorectal Carcinoma: An Aggressive Adenocarcinoma with Reduced CDX2 and Increased Cytokeratin 7 Immunohistochemical Expression. Human Pathology 45 (8):1704–12. 2014/08
[Full Text Article] [Pubmed: 24908142] -
Lao, Victoria Valinluck et al. Altered RECQ Helicase Expression in Sporadic Primary Colorectal Cancers. Translational Oncology 6 (4):458–69. 2013/08
[Pubmed: 23908689] [PMC: PMC3730021] -
Larraguibel, Jahdiel et al. Wnt Ligand-Dependent Activation of the Negative Feedback Regulator Nkd1. Molecular Biology of the Cell 26 (12):2375–84. 6/15/2015
[Full Text Article] [Pubmed: 25904337] [PMC: PMC4462952] -
Li, Gang et al. Altered Expression of Polycomb Group Genes in Glioblastoma Multiforme. PLoS ONE 8 (11). 11/15/2013
[Full Text Article] [Pubmed: 24260522] [PMC: PMC3829908] -
Li, Jing et al. PTEN, a Putative Protein Tyrosine Phosphatase Gene Mutated in Human Brain, Breast, and Prostate Cancer. Science 275 (5308):1943–47. 3/28/1997
[Full Text Article] [Pubmed: 9072974] -
Li, Ying et al. Special AT-Rich Sequence-Binding Protein 2 Acts as a Negative Regulator of Stemness in Colorectal Cancer Cells. World Journal of Gastroenterology 22 (38):8528–39. 10/14/2016
[Full Text Article] [Pubmed: 27784965] [PMC: PMC5064034] -
Liccardo, Raffaella et al. Novel Implications in Molecular Diagnosis of Lynch Syndrome. Gastroenterology Research and Practice 2017:2595098. 2017
[Full Text Article] [Pubmed: 28250766] [PMC: PMC5303590] -
Lin, Fan et al. Cadherin-17 and SATB2 Are Sensitive and Specific Immunomarkers for Medullary Carcinoma of the Large Intestine. Archives of Pathology & Laboratory Medicine 138 (8):1015–26. 1/17/2014
[Full Text Article] -
Lundberg, Ida V. et al. SOX2 Expression Is Associated with a Cancer Stem Cell State and Down-Regulation of CDX2 in Colorectal Cancer. BMC Cancer 16 (July):471. 7/13/2016
[Full Text Article] [Pubmed: 27411517] [PMC: PMC4944515] -
Merlos-Suárez, Anna et al. The Intestinal Stem Cell Signature Identifies Colorectal Cancer Stem Cells and Predicts Disease Relapse. Cell Stem Cell 8 (5):511–24. 5/6/2011
[Full Text Article] -
Mo, Dongliang et al. Human Helicase RECQL4 Drives Cisplatin Resistance in Gastric Cancer by Activating an AKT-YB1-MDR1 Signaling Pathway. Cancer Research 76 (10):3057–66. 5/15/2016
[Full Text Article] [Pubmed: 27013200] -
Moehle, Christoph et al. Aberrant Intestinal Expression and Allelic Variants of Mucin Genes Associated with Inflammatory Bowel Disease. Journal of Molecular Medicine 84 (12):1055–66. 12/1/2006
[Full Text Article] -
Moll, R. et al. Identification of Protein IT of the Intestinal Cytoskeleton as a Novel Type I Cytokeratin with Unusual Properties and Expression Patterns. The Journal of Cell Biology 111 (2):567–80. 1990/08
[Pubmed: 1696264] [PMC: PMC2116178] -
Morgan, R. L. et al. MOC-31 Aids in the Differentiation between Adenocarcinoma and Reactive Mesothelial Cells. Cancer 87 (6):390–94. 12/25/1999
[Pubmed: 10603193] -
Mouradov, Dmitri et al. Colorectal Cancer Cell Lines Are Representative Models of the Main Molecular Subtypes of Primary Cancer. Cancer Research 74 (12):3238–47. 6/15/2014
[Full Text Article] [Pubmed: 24755471] -
Nome, Torfinn et al. High Frequency of Fusion Transcripts Involving TCF7L2 in Colorectal Cancer: Novel Fusion Partner and Splice Variants. PLOS ONE 9 (3):e91264. 3/7/2014
[Full Text Article] -
Novellasdemunt, Laura et al. Targeting Wnt Signaling in Colorectal Cancer. A Review in the Theme: Cell Signaling: Proteins, Pathways and Mechanisms. American Journal of Physiology - Cell Physiology 309 (8):C511–21. 10/15/2015
[Full Text Article] [Pubmed: 26289750] [PMC: PMC4609654] -
Ollikainen, M. et al. Mechanisms of Inactivation of MLH1> in Hereditary Nonpolyposis Colorectal Carcinoma: A Novel Approach. Oncogene 26 (31):4541. 2007/07
[Full Text Article] -
Panarelli, Nicole C. et al. Tissue-Specific Cadherin CDH17 Is a Useful Marker of Gastrointestinal Adenocarcinomas With Higher Sensitivity Than CDX2. American Journal of Clinical Pathology 138 (2):211–22. 8/1/2012
[Full Text Article] -
Papadopoulos, N. et al. Mutation of a MutL Homolog in Hereditary Colon Cancer. Science 263 (5153):1625–29. 3/18/1994
[Full Text Article] [Pubmed: 8128251] -
Pezzolesi, Marcus G. et al. Comparative Genomic and Functional Analyses Reveal a Novel Cis-Acting PTEN Regulatory Element as a Highly Conserved Functional E-Box Motif Deleted in Cowden Syndrome. Human Molecular Genetics 16 (9):1058–71. 5/1/2007
[Full Text Article] -
Pino, Maria S. et al. THE CHROMOSOMAL INSTABILITY PATHWAY IN COLON CANCER. Gastroenterology 138 (6):2059–72. 2010/06
[Full Text Article] [Pubmed: 20420946] [PMC: PMC4243705] -
Platet, Nadine et al. The Tumor Suppressor CDX2 Opposes Pro-Metastatic Biomechanical Modifications of Colon Cancer Cells through Organization of the Actin Cytoskeleton. Cancer Letters 386 (February):57–64. 2/1/2017
[Full Text Article] [Pubmed: 27816490] -
Polat, E. et al. Diagnostic Value of Preoperative Serum Carcinoembryonic Antigen and Carbohydrate Antigen 19-9 in Colorectal Cancer. Current Oncology (Toronto, Ont.) 21 (1):e1-7. 2014/02
[Full Text Article] [Pubmed: 24523606] [PMC: PMC3921033] -
Rageul, Julie et al. KLF4-Dependent, PPARγ-Induced Expression of GPA33 in Colon Cancer Cell Lines. International Journal of Cancer. Journal International Du Cancer 125 (12):2802–9. 12/15/2009
[Full Text Article] [Pubmed: 19551868] [PMC: PMC2791338] -
Rhee, Ye-Young et al. CpG Island Methylator Phenotype-High Colorectal Cancers and Their Prognostic Implications and Relationships with the Serrated Neoplasia Pathway. Gut and Liver 11 (1):38–46. 2017/01
[Full Text Article] [Pubmed: 27885175] [PMC: PMC5221859] -
Rosty, Christophe et al. Germline Mutations in PMS2 and MLH1 in Individuals with Solitary Loss of PMS2 Expression in Colorectal Carcinomas from the Colon Cancer Family Registry Cohort. BMJ Open 6 (2):e010293. 2/1/2016
[Full Text Article] [Pubmed: 26895986] -
Rosty, Christophe et al. PIK3CA Activating Mutation in Colorectal Carcinoma: Associations with Molecular Features and Survival. PLOS ONE 8 (6):e65479. 6/13/2013
[Full Text Article] -
Sainz, Juan et al. Effect of Type 2 Diabetes Predisposing Genetic Variants on Colorectal Cancer Risk. The Journal of Clinical Endocrinology and Metabolism 97 (5):E845-851. 2012/05
[Full Text Article] [Pubmed: 22419714] -
Segditsas, S. et al. Colorectal Cancer and Genetic Alterations in the Wnt Pathway. Oncogene 25 (57):7531. 2006/12
[Full Text Article] -
Shah, Sandeep N. et al. Defective Mismatch Repair, Microsatellite Mutation Bias, and Variability in Clinical Cancer Phenotypes. Cancer Research 70 (2):431–35. 1/15/2010
[Full Text Article] [Pubmed: 20068152] [PMC: PMC2807994] -
Shimokawa, Mariko et al. Visualization and Targeting of LGR5+ Human Colon Cancer Stem Cells. Nature 545 (7653):187. 3/29/2017
[Full Text Article] -
Siegel, Rebecca L. et al. Colorectal Cancer Statistics, 2017. CA: A Cancer Journal for Clinicians 67 (3):177–93. 5/6/2017
[Full Text Article] [Pubmed: 28248415] -
Stolze, Britta et al. Comparative Analysis of KRAS Codon 12, 13, 18, 61, and 117 Mutations Using Human MCF10A Isogenic Cell Lines. Scientific Reports 5 (February):8535. 2/23/2015
[Full Text Article] -
Takahashi, Kazutoshi et al. Induction of Pluripotent Stem Cells from Mouse Embryonic and Adult Fibroblast Cultures by Defined Factors. Cell 126 (4):663–76. 8/25/2006
[Full Text Article] [Pubmed: 16904174] -
Takeishi, S. et al. Role of Fbxw7 in the Maintenance of Normal Stem Cells and Cancer-Initiating Cells. British Journal of Cancer 111 (6):1054. 2014/09
[Full Text Article] -
Velcich, Anna et al. Colorectal Cancer in Mice Genetically Deficient in the Mucin Muc2. Science 295 (5560):1726–29. 3/1/2002
[Full Text Article] [Pubmed: 11872843] -
Walsh, Michael D. et al. Expression of MUC2, MUC5AC, MUC5B, and MUC6 Mucins in Colorectal Cancers and Their Association with the CpG Island Methylator Phenotype. Modern Pathology 26 (12):1642. 2013/12
[Full Text Article] -
Win, Aung Ko et al. Prevalence and Penetrance of Major Genes and Polygenes for Colorectal Cancer. Cancer Epidemiology, Biomarkers & Prevention : A Publication of the American Association for Cancer Research, Cosponsored by the American Society of Preventive Oncology 26 (3):404–12. 2017/03
[Full Text Article] [Pubmed: 27799157] [PMC: PMC5336409] -
Worthley, Daniel L. et al. Familial Mutations in PMS2 Can Cause Autosomal Dominant Hereditary Nonpolyposis Colorectal Cancer. Gastroenterology 128 (5):1431–36. 2005/05
[Pubmed: 15887124] -
Wu, Julie et al. Distribution of BRAF T1799A(V600E) Mutations across Various Types of Benign Nevi: Implications for Melanocytic Tumorigenesis. The American Journal of Dermatopathology 29 (6):534–37. 2007/12
[Full Text Article] [Pubmed: 18032947] -
Xu, Jian Ming et al. PIK3CA Mutations Contribute to Acquired Cetuximab Resistance in Metastatic Colorectal Cancer Patients. Clinical Cancer Research, January, clincanres.2738.2016. 1/1/2017
[Full Text Article] [Pubmed: 28424201] -
Zhao, Jia-Xing et al. Serum CA19-9 as a Marker of Circulating Tumor Cells in First Reflux Blood of Colorectal Cancer Patients. Oncotarget 8 (40):67918–32. 9/15/2017
[Full Text Article] [Pubmed: 28978084] [PMC: PMC5620224] -
Zhu, Zi-yuan et al. Expression of Chromobox Homolog 2 in Colorectal Cancer and Its Clinical Significance. TUMOR 37 (10):1056–62. 10/25/2017
-
Comprehensive Molecular Characterization of Human Colon and Rectal Cancer. 2012. Nature 487 (7407):330–37. 7/18/2012
[Full Text Article] [Pubmed: 22810696] [PMC: PMC3401966]