Login
Registration enables users to use special features of this website, such as past
order histories, retained contact details for faster checkout, review submissions, and special promotions.
order histories, retained contact details for faster checkout, review submissions, and special promotions.
Forgot password?
Registration enables users to use special features of this website, such as past
order histories, retained contact details for faster checkout, review submissions, and special promotions.
order histories, retained contact details for faster checkout, review submissions, and special promotions.
Quick Order
Products
Antibodies
ELISA and Assay Kits
Research Areas
Infectious Disease
Resources
Purchasing
Reference Material
Contact Us
Location
Corporate Headquarters
Vector Laboratories, Inc.
6737 Mowry Ave
Newark, CA 94560
United States
Telephone Numbers
Customer Service: (800) 227-6666 / (650) 697-3600
Contact Us
Additional Contact Details
Login
Registration enables users to use special features of this website, such as past
order histories, retained contact details for faster checkout, review submissions, and special promotions.
order histories, retained contact details for faster checkout, review submissions, and special promotions.
Forgot password?
Registration enables users to use special features of this website, such as past
order histories, retained contact details for faster checkout, review submissions, and special promotions.
order histories, retained contact details for faster checkout, review submissions, and special promotions.
Quick Order
Lung Cancer Immunohistochemistry Markers
Lung cancer is the second most common cause of cancer deaths in the U.S. Lung cancers exist as three subtypes: non-small cell lung cancers (NSCLC),
which are comprised of the adenocarcinoma and squamous cell carcinoma subtypes that together account for 75% of cases, and small cell lung cancers (neuroendocrine origin)
which account for the remainder.
The availability of targeted therapies makes it essential to correctly subtype NSCLCs: IHC markers such as
NKX2-1 / TTF-1, Napsin A,
and surfactant A (adenocarcinoma) or p63,
CK5 / CK6,
SOX2 and Desmoglein-3
(squamous cell carcinoma) can be used to identify the subtype in cases where the morphology is unclear, or with biopsies where there is
insufficient material to make the diagnosis (Noh, 2012; Gurda, 2015). Small cell lung cancers can be identified with markers such as
LMWK,
CAM5.2,
chromogranin, synaptophysin,
CD56, and NKX2-1 / TTF-1 (Travis, 2012).
Although platinum-based chemotherapy has been the standard treatment for metastatic NSCLC, lung cancers with mutations associated with epidermal growth factor receptor
(EGFR) and anaplastic lymphoma kinase (ALK) may respond to specific kinase inhibitors, so there is a
need to identify the subset that show these mutations (Cooper, 2011; Malhotra, 2017; Kumar, Curr Probl Cancer 2017 Mar - Apr;41(2):111-124). Furthermore...
Click to Expand Text ▸
Primary IHC Markers In Lung Cancer
PDL1 (CD274)
PDL1 (CD274), or programmed cell death-ligand 1, is an immune checkpoint receptor involved in immune escape in cancer, and it is upregulated in many cancers including NSCLC (Zhang, 2016). High levels of expression of PDL1 are associated with poor prognosis in NSCLC and pulmonary lymphoepithelioma-like carcinoma (Zhang 2016). Expression is also associated with EGFR mutations, the onset of which may directly lead to upregulation of PDL1 (Chen, 2015; Tang, 2015). Levels of expression of PDL1 detected through immunohistochemistry can predict response to PD1 therapy (Sorensen, 2016). Current clinical trials involving PDL1 inhibitors have shown impressive results in treating advanced stage cancers, particularly melanoma. Placental trophoblasts and macrophages are the best positive controls for this target.
Staining:
This protein may have cytoplasmic or membranous staining in immune cells and tumor cells.
LSBio's recommended antibody to PDL1 / CD274 for use in immunohistochemistry is PDL1 / CD274 Antibody LS‑B10562.
Click here for specifications and further information on this antibody.
Click here for specifications and further information on this antibody.
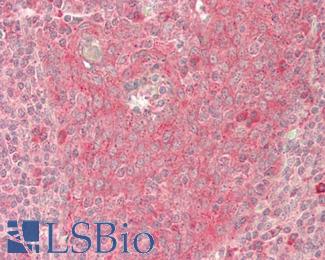
 Anti-B7-H1 / PD-L1 / CD274 antibody IHC staining of human testis. Immunohistochemistry of formalin-fixed, paraffin-embedded tissue after heat-induced antigen retrieval. Antibody LS-B10562 dilution 1:100.
Anti-B7-H1 / PD-L1 / CD274 antibody IHC staining of human testis. Immunohistochemistry of formalin-fixed, paraffin-embedded tissue after heat-induced antigen retrieval. Antibody LS-B10562 dilution 1:100.
PDL1 / CD274 Antibody LS-B10562
PD1 (PDCD1 / CD279)
PD1 (Programmed Death Receptor 1) is an immune checkpoint protein active in T cells that is a target alongside its ligand PDL1 and also CTLA-4 for immunotherapy in lung and other cancers (Alsaab, 2017). PD1 is involved in negatively regulating T cell inflammatory activity and, when bound to receptors on tumor cells, can work to subdue tumor suppression by inhibiting the immune response. Targeted inhibition of PD1 itself can therefore function as an anti-cancer therapy by reactivating this response (Jin, 2011; Francisco, 2010; Fife, 2011).
Staining:
PD1 is expected to have membranous staining in immune cells and tumor cells.
LSBio's recommended antibody to PD1 (PDCD1) for use in immunohistochemistry is PD1 (PDCD1) Antibody LS‑B7883.
Click here for specifications and further information on this antibody.
Click here for specifications and further information on this antibody.
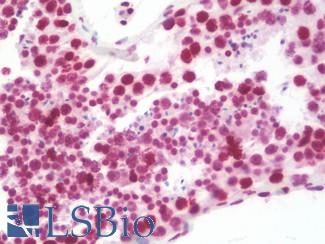
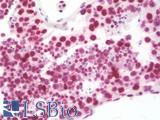
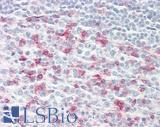 Anti-CD279 / PD-1 antibody IHC of human tonsil. Immunohistochemistry of formalin-fixed, paraffin-embedded tissue after heat-induced antigen retrieval. Antibody LS-B7883 dilution 20 ug/ml.
Anti-CD279 / PD-1 antibody IHC of human tonsil. Immunohistochemistry of formalin-fixed, paraffin-embedded tissue after heat-induced antigen retrieval. Antibody LS-B7883 dilution 20 ug/ml.
PD1 (PDCD1) Antibody LS-B7883
AHR
Aryl hydrocarbon receptor (AHR) is a transcription factor involved in xenobiotic metabolism and autophagy and it is expressed in many tissues including lung. In cancer, AHR is involved in migration and proliferation, and is upregulated in pancreatic, liver, head and neck, lung, thyroid and gastrointestinal tumors. AHR appears to function in both tumor suppression and tumorigenesis depending on the cell type and tissue (Tsai, 2017; Safe, 2013). In the lung, AHR expression is correlated with smoking-induced tumors, where carcinogens such as BaP directly bind to AHR and induce xenobiotic metabolism. Activation of AHR leads to upregulation of CYP1A1 and other cytochrome P450 enzymes, which are responsible for metabolizing carcinogens but can sometimes create a more damaging substance in error (Tsay, 2013). Higher expression of AHR in lung carcinomas is associated with a worse prognosis, particularly in tumors with mutant EGFR. In these tumors, the activation of AHR induces oncogenic Src expression and functional proliferative signaling (PI3K/Akt), resulting in resistance to tyrosine kinase inhibitors that target EGFR (Ye, 2017).
Staining:
AHR is expected to have nuclear and cytoplasmic staining.
LSBio's recommended antibody to AHR for use in immunohistochemistry is AHR Antibody LS‑A3018.
Click here for specifications and further information on this antibody.
Click here for specifications and further information on this antibody.
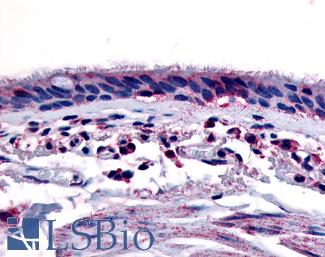
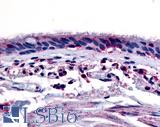 Anti-Aryl Hydrocarbon Receptor antibody LS-A3018 IHC of human respiratory epithelium and bronchial smooth muscle. Immunohistochemistry of formalin-fixed, paraffin-embedded tissue after heat-induced antigen retrieval.
Anti-Aryl Hydrocarbon Receptor antibody LS-A3018 IHC of human respiratory epithelium and bronchial smooth muscle. Immunohistochemistry of formalin-fixed, paraffin-embedded tissue after heat-induced antigen retrieval.
AHR Antibody LS-A3018
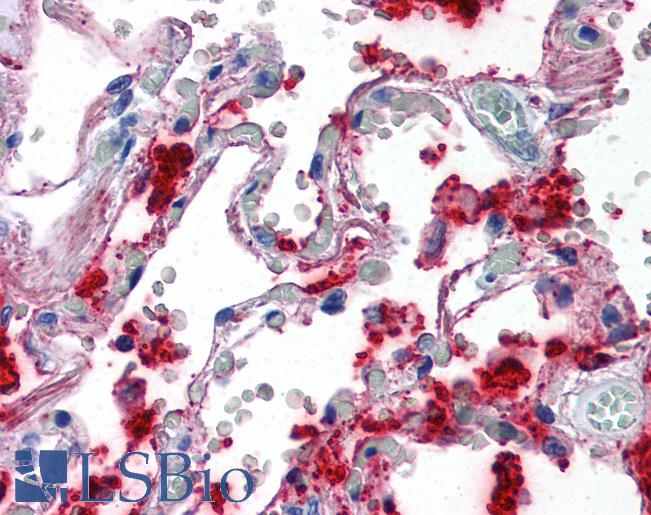
Surfactant Protein A (SFTPA1)
Surfactant protein A (SFTPA1) is involved in host defense in the lung and its expression is necessary for tubular myelin formation and in reducing surface tension within alveoli (Lopez-Rodriguez, 2016). Germline mutations in SFTPA1 that result in impaired surfactant secretion and respiratory insufficiency have been implicated in a hereditary predisposition for lung fibrosis and cancer (Nathan, 2016). The specific nature of surfactant A to lung tissue has made it a useful marker in lung diseases including adenocarcinoma (Takahashi, 2006). It has comparable sensitivity to marker NKX2-1 /TTF-1 and is particularly useful in distinguishing NKX2-1-positive lung from thyroid tumors (Tan, 2008).
Staining:
Staining of SFTPA1 is expected to be cytoplasmic in lung tissue pneumocytes.
LSBio's recommended antibody to Surfactant Protein A for use in immunohistochemistry is Surfactant Protein A Antibody LS‑B2543.
Click here for specifications and further information on this antibody.
Click here for specifications and further information on this antibody.
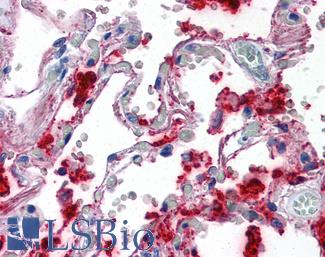
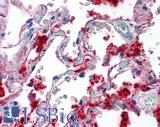 Anti-SFTPA1 / Surfactant Protein A antibody IHC of human lung. Immunohistochemistry of formalin-fixed, paraffin-embedded tissue after heat-induced antigen retrieval. Antibody LS-B2543 concentration 10 ug/ml.
Anti-SFTPA1 / Surfactant Protein A antibody IHC of human lung. Immunohistochemistry of formalin-fixed, paraffin-embedded tissue after heat-induced antigen retrieval. Antibody LS-B2543 concentration 10 ug/ml.
Surfactant Protein A Antibody LS-B2543
SOX2
SOX2 is a Yamanaka factor and master regulator involved in embryogenesis and transdifferentiation (Takahashi, 2006; Whyte, 2013). In squamous cell carcinoma of the lung, SOX2 frequently has amplified copy number (~20% of cases) and overexpression of the gene is found in roughly 90% of cases (Fukazawa, 2015; Rudin, 2012). Upregulation of SOX2 is likely a primary determinant of the transformation of basal cells into carcinomas of various subtypes, and the variable upstream inactivation of different tumor suppressors seems to decide which subtype. Specifically, in the lung SOX2 overexpression results in squamous cell carcinoma following loss of PTEN, CDKN2A/CDKN2B and/or STK11, while upregulation subsequent to TP53 inactivation results in adenocarcinoma (Murray, 2016; Ferone, 2016).
Staining:
Staining for this protein is expected to be nuclear.
LSBio's recommended antibody to SOX2 for use in immunohistochemistry is SOX2 Antibody LS‑B4562.
Click here for specifications and further information on this antibody.
Click here for specifications and further information on this antibody.
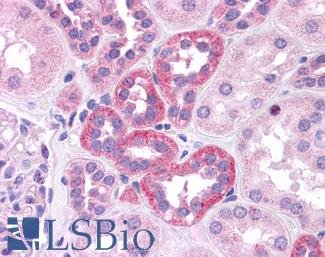
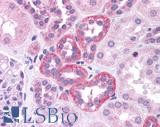 Anti-SOX2 antibody IHC of human kidney. Immunohistochemistry of formalin-fixed, paraffin-embedded tissue after heat-induced antigen retrieval. Antibody LS-B4562 concentration 5 ug/ml.
Anti-SOX2 antibody IHC of human kidney. Immunohistochemistry of formalin-fixed, paraffin-embedded tissue after heat-induced antigen retrieval. Antibody LS-B4562 concentration 5 ug/ml.
SOX2 Antibody LS-B4562
APOBEC3B
APOBEC3B is a cytidine deaminase involved in DNA repair and also somatic hypermutation. The deregulation of APOBEC family members including APOBEC3B has been implicated as a frequent driving source of mutagenesis in breast cancer and in other tissues (Nik-Zainal, 2012; McBride, 2008; Hwang, 2015). Campbell et al have also identified APOBEC mutation signatures in lung cancer, and further studies have found evidence of APOBEC3B upregulation in lung cancer and a correlation between increased expression and worse prognosis in NSCLC , making it an interesting and potentially important target for immunohistochemistry (Campbell 2016; Sasaki, 2014; Shumei, 2016).
Staining:
Staining of APOBEC3B is expected to be nuclear.
LSBio's recommended antibody to APOBEC3B for use in immunohistochemistry is APOBEC3B Antibody LS‑B12051.
Click here for specifications and further information on this antibody.
Click here for specifications and further information on this antibody.
MET
MET (hepatocyte growth factor receptor / HGFR) is a receptor tyrosine kinase that is normally active in embryonic stem cells and progenitor cells. During embryogenesis, MET expression allows cells to burrow into adjacent areas in order to generate new embryonic structures, and in adult tissues, this invasive potential is a normal and essential component of wound repair. MET also promotes cell growth and inhibits apoptosis. In cancers, Met activation is associated with an aggressive, invasive phenotype and confers a poor prognosis, triggering tumor growth and ingrowth of new blood vessels. MET deregulation is common in human cancers, and contributes to therapeutic resistance, as it can be induced by chemotherapeutic agents and radiation. Hence Met inhibition, either targeting the receptor or HGFR ligand, is an area of interest, both as a primary and adjuvant means of treating cancer.
Staining:
MET is expected to have membranous and cytoplasmic staining.
LSBio's recommended antibody to c-MET for use in immunohistochemistry is c-MET Antibody LS‑B2812.
Click here for specifications and further information on this antibody.
Click here for specifications and further information on this antibody.
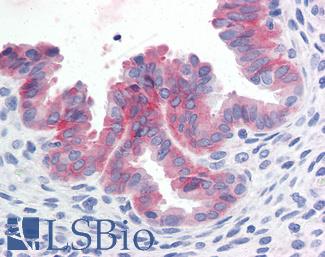
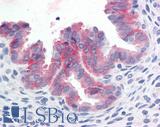 Anti-c-Met antibody IHC of human uterus. Immunohistochemistry of formalin-fixed, paraffin-embedded tissue after heat-induced antigen retrieval. Antibody LS-B2812 concentration 10 ug/ml.
Anti-c-Met antibody IHC of human uterus. Immunohistochemistry of formalin-fixed, paraffin-embedded tissue after heat-induced antigen retrieval. Antibody LS-B2812 concentration 10 ug/ml.
c-MET Antibody LS-B2812
Cell-Type Specific Markers In Lung Cancer
CK5 (KRT5)
CK5 (KRT5) is positive in mesothelioma and in squamous carcinoma and rare in adenocarcinoma. It is a useful marker when used alongside CK6 and TP63 and in comparison with NKX2-1 staining for subtyping squamous cell versus adenocarcinomas of the lung (Carney, 2015; Rekhtman, 2011)
Staining:
Staining may be membranous and cytoplasmic.
LSBio's recommended antibody to KRT5 / CK5 for use in immunohistochemistry is KRT5 / CK5 Antibody LS‑B9009.
Click here for specifications and further information on this antibody.
Click here for specifications and further information on this antibody.
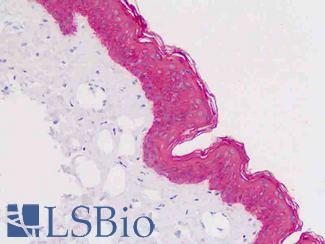
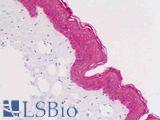 Anti-KRT5 / CK5 / Cytokeratin 5 antibody IHC staining of human skin, epidermis. Immunohistochemistry of formalin-fixed, paraffin-embedded tissue after heat-induced antigen retrieval.
Anti-KRT5 / CK5 / Cytokeratin 5 antibody IHC staining of human skin, epidermis. Immunohistochemistry of formalin-fixed, paraffin-embedded tissue after heat-induced antigen retrieval.
KRT5 / CK5 Antibody LS-B9009
CK6 (KRT6)
CK6 (KRT6) stains positively in mesothelioma and squamous carcinoma and is rare in adenocarcinoma. KRT6 is useful as a marker when used together with TP63 and KRT5 for subtyping squamous cell versus adenocarcinomas of the lung (Carney, 2015; Rekhtman, 2011).
Staining:
Staining is expected to be cytoplasmic.
LSBio's recommended antibody to KRT6 / CK6 for use in immunohistochemistry is KRT6 / CK6 Antibody LS‑C392262.
Click here for specifications and further information on this antibody.
Click here for specifications and further information on this antibody.
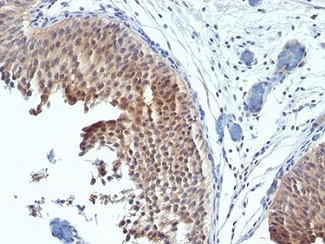
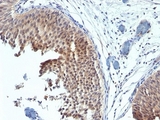 Formalin-fixed, paraffin-embedded human bladder carcinoma stained with Cytokeratin 6 antibody (SPM269). This image was taken for the unmodified form of this product. Other forms have not been tested.
Formalin-fixed, paraffin-embedded human bladder carcinoma stained with Cytokeratin 6 antibody (SPM269). This image was taken for the unmodified form of this product. Other forms have not been tested.
KRT6 / CK6 Antibody LS-C392262
Chromogranin A (CHGA)
CHGA (Chromogranin A) encodes a protein involved in the production of secretory vesicles in endocrine and neuroendocrine cells. Levels of this protein correlate with the number of secretory vesicles present, and staining of this protein is useful for subtyping neuroendocrine tumors in multiple tissues including the lung (Gut, 2016; Syversen, 2004). Serum of patients with small cell lung cancer can be checked for circulating CHGA, and in this context it has been shown to have more sensitivity than neuroendocrine marker NSE (Taneja, 2004). Sensitivity of CHGA is expected to increase with clinical stage (Seregni, 2001).
Staining:
Staining for Chromogranin A is expected to be cytoplasmic.
LSBio's recommended antibody to Chromogranin A for use in immunohistochemistry is Chromogranin A Antibody LS‑B6265.
Click here for specifications and further information on this antibody.
Click here for specifications and further information on this antibody.
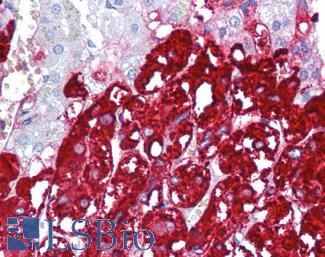
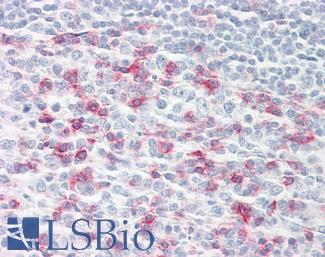
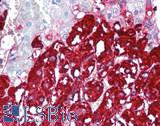 Anti-CHGA / Chromogranin A antibody IHC of human adrenal. Immunohistochemistry of formalin-fixed, paraffin-embedded tissue after heat-induced antigen retrieval. Antibody LS-B6265 dilution 1:200.
Anti-CHGA / Chromogranin A antibody IHC of human adrenal. Immunohistochemistry of formalin-fixed, paraffin-embedded tissue after heat-induced antigen retrieval. Antibody LS-B6265 dilution 1:200.
Chromogranin A Antibody LS-B6265
NKX2-1 (TTF-1)
NKX2-1 (also known as TTF-1) is a transcription factor with a necessary function in the differentiation of lung, thyroid and forebrain cells, and it regulates the expression of surfactant proteins in the lung that are required for host defense and stability (Boggaram, 2009) . NKX2-1 is a useful cancer marker for distinguishing a primary adenocarcinoma from other metastases, particularly when combined with Napsin A in a marker panel (Anagnostou, 2008; Ye, 2011). It is often expressed in adenocarcinoma, where expression is correlated with EGFR mutation and also higher rates of survival in some patients (Wan, 2014; Anagnostou, 2008).
Staining:
NKX2-1 is expected to have nuclear staining.
LSBio's recommended antibody to NKX2-1 / TTF-1 for use in immunohistochemistry is NKX2-1 / TTF-1 Antibody LS‑B10666.
Click here for specifications and further information on this antibody.
Click here for specifications and further information on this antibody.
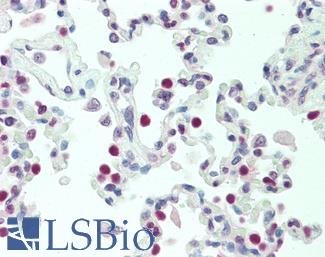
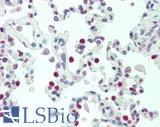 Anti-TTF1 / TTF-1 / NKX2-1 antibody IHC staining of human lung. Immunohistochemistry of formalin-fixed, paraffin-embedded tissue after heat-induced antigen retrieval. Antibody LS-B10666 dilution 1:100.
Anti-TTF1 / TTF-1 / NKX2-1 antibody IHC staining of human lung. Immunohistochemistry of formalin-fixed, paraffin-embedded tissue after heat-induced antigen retrieval. Antibody LS-B10666 dilution 1:100.
NKX2-1 / TTF-1 Antibody LS-B10666
CD56 (NCAM1)
CD56 (NCAM1 / neural cell adhesion molecule 1) is a cell adhesion protein expressed in natural killer cells where its expression acts as a reliable marker of activation (Pruitt, 2011; Van Acker, 2017). It is often considered the most specific and sensitive of small cell neuroendocrine markers in the lung (Rekhtman, 2010), and is particularly useful in diagnosing small cell cancers with intense crush artefact (Kontogianni, 2005).
Staining:
Staining is expected to be primarily cytoplasmic.
LSBio's recommended antibody to NCAM1 / CD56 for use in immunohistochemistry is NCAM1 / CD56 Antibody LS‑B5569.
Click here for specifications and further information on this antibody.
Click here for specifications and further information on this antibody.
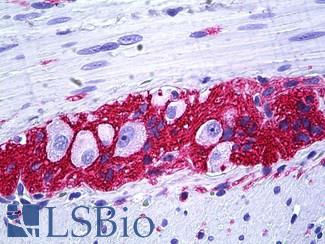
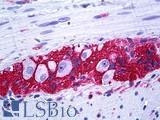 Anti-CD56 antibody IHC of human intestine, ganglion cells. Immunohistochemistry of formalin-fixed, paraffin-embedded tissue after heat-induced antigen retrieval. Antibody LS-B5569 dilution 1:100.
Anti-CD56 antibody IHC of human intestine, ganglion cells. Immunohistochemistry of formalin-fixed, paraffin-embedded tissue after heat-induced antigen retrieval. Antibody LS-B5569 dilution 1:100.
NCAM1 / CD56 Antibody LS-B5569
DSG3
DSG3 (Desmoglein-3) identifies squamous differentiaton and is a highly specific and sensitive marker for pulmonary squamous cell carcinomas, particularly in subtyping from other classes of lung cancer (Savci-Heijink, 2009). Desmosomal protein is expressed in squamous epithelium where it forms part of the desmosomal structures that comprise cell junctions. Upregulation of DSG3 has been correlated with metastasis in a number of cancers including lung, potentially due to DSG3’s role in cell-adhesion and migration mechanics via regulation of the WNT/B-Catenin/TCF signaling pathway (Weed, 2015).
Staining:
Staining for DSG3 is expected to be membranous.
LSBio's recommended antibody to DSG3 for use in immunohistochemistry is DSG3 Antibody LS‑B14031.
Click here for specifications and further information on this antibody.
Click here for specifications and further information on this antibody.
Synaptophysin (SYP)
SYP (Synaptophysin) is a protein associated with neuronal synaptic vesical membranes that regulates endocytosis. In immunohistochemistry, Synaptophysin acts as a small cell neuroendocrine marker, and it is a very sensitive marker of neuroendocrine differentiation (Wiedenmann, 1986; Fisseler-Eckhoff, 2012).
Staining:
Staining for SYP is expected to be cytoplasmic in neuroendocrine cells.
LSBio's recommended antibody to Synaptophysin for use in immunohistochemistry is Synaptophysin Antibody LS‑B3393.
Click here for specifications and further information on this antibody.
Click here for specifications and further information on this antibody.
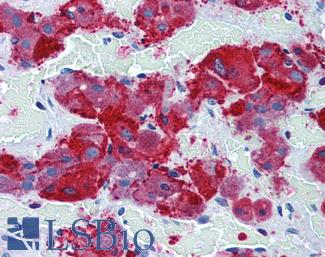
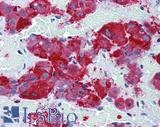 Anti-SYP / Synaptophysin antibody IHC of human adrenal. Immunohistochemistry of formalin-fixed, paraffin-embedded tissue after heat-induced antigen retrieval. Antibody LS-B3393 dilution 1:50.
Anti-SYP / Synaptophysin antibody IHC of human adrenal. Immunohistochemistry of formalin-fixed, paraffin-embedded tissue after heat-induced antigen retrieval. Antibody LS-B3393 dilution 1:50.
Synaptophysin Antibody LS-B3393
Napsin A
Napsin A (NAPSA) is a secreted pepsidase produced in type II pneumocytes and renal tubular epithelium. It is involved in processing surfactant B in the lung and acts as a useful cytoplasmic marker for primary lung adenocarcinomas (Brasch, 2003; Ueno, 2004; Siddiqui, 2012). When paired with transcription factor TTF-1 and squamous markers TP63 and KRT5 in a marker panel, Napsin A is useful for distinguishing primary lung adenocarcinomas from other metastases (Ye, 2011; Turner, 2012; Siddiqui, 2012 ).
Staining:
Staining for NAPSA is expected to be cytoplasmic.
LSBio's recommended antibody to NAPSA / Napsin A for use in immunohistochemistry is NAPSA / Napsin A Antibody LS‑C142870.
Click here for specifications and further information on this antibody.
Click here for specifications and further information on this antibody.
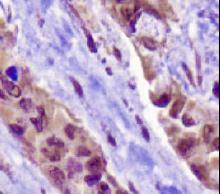
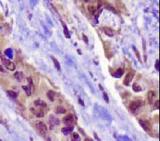 NAPSIN1 Antibody (TMU-Ad02) - IHC of Napsin1 on human lung adenocarcinoma. This image was taken for the unconjugated form of this product. Other forms have not been tested.
NAPSIN1 Antibody (TMU-Ad02) - IHC of Napsin1 on human lung adenocarcinoma. This image was taken for the unconjugated form of this product. Other forms have not been tested.
NAPSA / Napsin A Antibody LS-C142870
Oncogenic Markers (Wild Type) In Lung Cancer
EGFR
EGFR (Epidermal growth factor receptor) is an ERBB family receptor tyrosine kinase involved in regulating proliferation, apoptosis and other aspects of the cell cycle through the MAPK and PI3K/AKT signaling pathways. Mutations or overexpression of EGFR, often coinciding with increased copy number of the gene, occur in 40-90% of NSCLC tumors and mutations are recurrent in adenocarcinomas and in never-smokers (Bethune, 2010; Oronsky, 2017). As a biomarker, EGFR expression predicts response to tyrosine kinase inhibitor chemotherapy (TKI's) alongside testing for common activating mutations in EGFR (such as the L858R substitution) (Ladanyi, 2008). Individuals with inactivating mutations or with wild-type EGFR expression do not tend to see benefits from TKI treatment, and it is necessary to test for levels of wild type EGFR and mutation status to determine the appropriate therapy (Oronsky, 2017).
Staining:
EGFR is expected to have membranous and cytoplasmic staining in IHC.
LSBio's recommended antibody to EGFR for use in immunohistochemistry is EGFR Antibody LS‑B2199.
Click here for specifications and further information on this antibody.
Click here for specifications and further information on this antibody.
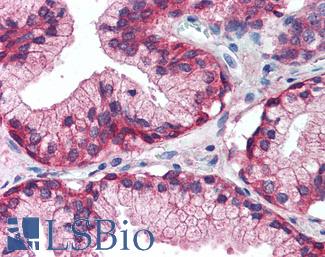
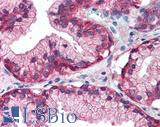 Anti-EGFR antibody IHC of human prostate. Immunohistochemistry of formalin-fixed, paraffin-embedded tissue after heat-induced antigen retrieval. Antibody LS-B2199 concentration 5 ug/ml.
Anti-EGFR antibody IHC of human prostate. Immunohistochemistry of formalin-fixed, paraffin-embedded tissue after heat-induced antigen retrieval. Antibody LS-B2199 concentration 5 ug/ml.
EGFR Antibody LS-B2199
KRAS
KRAS is a RAS/MAPK signaling pathway kinase with driving hotspot mutations in many tissues. KRAS is mutated in roughly 30% of lung adenocarcinomas and is less frequent in in squamous NSCLC (~5%), and it does not have detectable mutations in small cell lung cancer (Bhattacharya, 2015; Westcott, 2013). The most common KRAS mutation in lung cancer is the substitution G12C, but G12V and G12D also frequently occur (Bhattacharya, 2015).
Staining:
Staining is expected to be primarily cytoplasmic.
LSBio's recommended antibody to KRAS for use in immunohistochemistry is KRAS Antibody LS‑B4683.
Click here for specifications and further information on this antibody.
Click here for specifications and further information on this antibody.
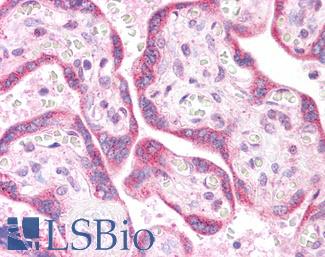
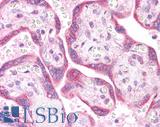 Anti-KRAS antibody IHC of human placenta. Immunohistochemistry of formalin-fixed, paraffin-embedded tissue after heat-induced antigen retrieval. Antibody LS-B4683 concentration 5 ug/ml. This image was taken for the unconjugated form of this product. Other forms have not been tested.
Anti-KRAS antibody IHC of human placenta. Immunohistochemistry of formalin-fixed, paraffin-embedded tissue after heat-induced antigen retrieval. Antibody LS-B4683 concentration 5 ug/ml. This image was taken for the unconjugated form of this product. Other forms have not been tested.
KRAS Antibody LS-B4683
References ▷
-
Anagnostou, Valsamo K. et al. Thyroid Transcription Factor 1 Is an Independent Prognostic Factor for Patients With Stage I Lung Adenocarcinoma.Journal of Clinical Oncology 1/10/2009; 27.2; 271-278
[Full Text Article] -
Bethune, Gillian et al. Epidermal growth factor receptor (EGFR) in lung cancer: an overview and update.Journal of Thoracic Disease 2010-03; 2.1; 48-51
[Pubmed: 22263017] [PMC: PMC3256436] -
Bhattacharya, Saveri et al. KRAS mutant lung cancer: progress thus far on an elusive therapeutic target.Clinical and Translational Medicine 12/14/2015; 4;
[Full Text Article] [Pubmed: 26668062] [PMC: PMC4678136] -
Boggaram, Vijay et al. Thyroid transcription factor-1 (TTF-1/Nkx2.1/TITF1) gene regulation in the lung.Clinical Science (London, England: 1979) 2009-01; 116.1; 27-35
[Full Text Article] [Pubmed: 19037882] -
Brasch, Frank et al. Involvement of Napsin A in the C- and N-terminal Processing of Surfactant Protein B in Type-II Pneumocytes of the Human Lung.Journal of Biological Chemistry 12/5/2003; 278.49; 49006-49014
[Full Text Article] [Pubmed: 13129928] -
Brown, Louise et al. Desmoglein 3: A Help or a Hindrance in Cancer Progression?.Cancers 1/26/2015; 7.1; 266-286
[Full Text Article] [Pubmed: 25629808] [PMC: PMC4381258] -
Campbell, Joshua D. et al. Distinct patterns of somatic genome alterations in lung adenocarcinomas and squamous cell carcinomas.Nature Genetics 2016; 48.6; 607-616
[Full Text Article] [Pubmed: 27158780] [PMC: PMC4884143] -
Carney, John M. et al. Immunostaining in Lung Cancer for the Clinician. Commonly Used Markers for Differentiating Primary and Metastatic Pulmonary Tumors.Annals of the American Thoracic Society 2/20/2015; 12.3; 429-435
[Full Text Article] -
Cerami, Ethan et al. The cBio Cancer Genomics Portal: An Open Platform for Exploring Multidimensional Cancer Genomics Data.Cancer Discovery 5/1/2012; 2.5; 401-404
[Full Text Article] [Pubmed: 22588877] -
Chen, Nan et al. Upregulation of PD-L1 by EGFR Activation Mediates the Immune Escape in EGFR-Driven NSCLC: Implication for Optional Immune Targeted Therapy for NSCLC Patients with EGFR Mutation.Journal of Thoracic Oncology 6/1/2015; 10.6; 910-923
[Full Text Article] -
Conde, Esther et al. The Use of P63 Immunohistochemistry for the Identification of Squamous Cell Carcinoma of the Lung.PLOS ONE 8/17/2010; 5.8; e12209
[Full Text Article] -
Cooper, Wendy A. et al. What's new in non-small cell lung cancer for pathologists: the importance of accurate subtyping, EGFR mutations and ALK rearrangements.Pathology 2011-02; 43.2; 103-115
[Full Text Article] [Pubmed: 21233671] -
Dacic, Sanja et al. ALK FISH patterns and the detection of ALK fusions by next generation sequencing in lung adenocarcinoma.Oncotarget 10/17/2016; 7.50; 82943-82952
[Full Text Article] [Pubmed: 27769042] [PMC: PMC5347743] -
El-Telbany, Ahmed et al. Cancer Genes in Lung Cancer.Genes & Cancer 2012-07; 3.43289; 467-480
[Full Text Article] [Pubmed: 23264847] [PMC: PMC3527990] -
Ferone, Giustina et al. SOX2 Is the Determining Oncogenic Switch in Promoting Lung Squamous Cell Carcinoma from Different Cells of Origin.Cancer Cell 10/10/2016; 30.4; 519-532
[Full Text Article] [Pubmed: 27728803] -
Fife, BT et al. The role of the PD-1 pathway in autoimmunity and peripheral tolerance. Ann N Y Acad Sci. 2011 Jan;1217:45-59.
[Full Text Article] [Pubmed: 21276005] -
Fisseler-Eckhoff, Annette et al. Neuroendocrine Tumors of the Lung.Cancers 7/31/2012; 4.3; 777-798
[Full Text Article] [Pubmed: 24213466] [PMC: PMC3712715] -
Francisco, et al. The PD-1 Pathway in Tolerance and Autoimmunity. Immunological reviews. 2010;236:219-242.
[Full Text Article] [Pubmed: 20636820] [PMC: PMC2919275] -
Fukazawa, Takuya et al. SOX2 suppresses CDKN1A to sustain growth of lung squamous cell carcinoma.Scientific Reports 2/5/2016; 6; 20113
[Full Text Article] -
Gao, Jianjiong et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal.Science Signaling 4/2/2013; 6.269; pl1
[Full Text Article] [Pubmed: 23550210] [PMC: PMC4160307] -
Gurda, Grzegorz T et al. Utility of five commonly used immunohistochemical markers TTF-1, Napsin A, CK7, CK5/6 and P63 in primary and metastatic adenocarcinoma and squamous cell carcinoma of the lung: a retrospective study of 246 fine needle aspiration cases.Clinical and Translational Medicine 4/21/2015; 4;
[Full Text Article] [Pubmed: 25977750] [PMC: PMC4417108] -
Gut, Paweł et al. Chromogranin A – unspecific neuroendocrine marker. Clinical utility and potential diagnostic pitfalls.Archives of Medical Science : AMS 2/1/2016; 12.1; 43109
[Full Text Article] [Pubmed: 26925113] [PMC: PMC4754364] -
Huang, Tao et al. Targeting histone methylation for colorectal cancer.Therapeutic Advances in Gastroenterology 2017-01; 10.1; 114-131
[Full Text Article] [Pubmed: 28286564] [PMC: PMC5330608] -
Hwang, Ki-Tae et al. Prognostic Influence of BCL2 on Molecular Subtypes of Breast Cancer.Journal of Breast Cancer 2017-03; 20.1; 54-64
[Full Text Article] [Pubmed: 28382095] [PMC: PMC5378580] -
Jin, HT et al. Role of PD-1 in regulating T-cell immunity. Curr Top Microbiol Immunol. 2011;350:17-37.
[Full Text Article] [Pubmed: 21061197] -
Kalhor, Neda et al. TTF-1 and p63 for distinguishing pulmonary small-cell carcinoma from poorly differentiated squamous cell carcinoma in previously pap-stained cytologic material.Modern Pathology 2006-08; 19.8; 1117
[Full Text Article] -
Kanwal, Madiha et al. Familial risk for lung cancer.Oncology Letters 2017-02; 13.2; 535-542
[Full Text Article] [Pubmed: 28356926] [PMC: PMC5351216] -
Kontogianni, K. et al. CD56: a useful tool for the diagnosis of small cell lung carcinomas on biopsies with extensive crush artefact.Journal of Clinical Pathology 2005-09; 58.9; 978-980
[Full Text Article] [Pubmed: 16126882] [PMC: PMC1770835] -
Ladanyi, Marc et al. Lung adenocarcinoma: guiding EGFR-targeted therapy and beyond.Modern Pathology 4/25/2008; 21.S2; S16
[Full Text Article] -
Lopez-Rodriguez, Elena et al. Human Pulmonary Surfactant Protein SP-A1 Provides Maximal Efficiency of Lung Interfacial Films.Biophysical Journal 8/9/2016; 111.3; 524-536
[Full Text Article] [Pubmed: 27508436] [PMC: PMC4982931] -
Malhotra, Jyoti et al. Current state of immunotherapy for non-small cell lung cancer.Translational Lung Cancer Research 2017-04; 6.2; 196-211
[Full Text Article] [Pubmed: 28529902] [PMC: PMC5420529] -
McBride, Kevin M. et al. Regulation of class switch recombination and somatic mutation by AID phosphorylation.The Journal of Experimental Medicine 10/27/2008; 205.11; 2585-2594
[Full Text Article] [Pubmed: 18838546] [PMC: PMC2571933] -
Morganella, Sandro et al. The topography of mutational processes in breast cancer genomes.Nature Communications 5/2/2016; 7; 11383
[Full Text Article] -
Murray, Nicole R. et al. SOX2 Determines Lineage Restriction: Modeling Lung Squamous Cell Carcinoma in the Mouse.Cancer Cell 10/10/2016; 30.4; 505-507
[Full Text Article] -
Nathan, Nadia et al. Germline SFTPA1 mutation in familial idiopathic interstitial pneumonia and lung cancer.Human Molecular Genetics 4/15/2016; 25.8; 1457-1467
[Full Text Article] [Pubmed: 26792177] -
Nik-Zainal, Serena et al. Landscape of somatic mutations in 560 breast cancer whole genome sequences.Nature 5/2/2016; 534.7605; 47-54
[Full Text Article] [Pubmed: 27135926] [PMC: PMC4910866] -
Nik-Zainal, Serena et al. Mutational processes molding the genomes of 21 breast cancers.Cell 5/25/2012; 149.5; 979-993
[Full Text Article] [Pubmed: 22608084] [PMC: PMC3414841] -
Nik-Zainal, Serena et al. Mutational Signatures in Breast Cancer: The Problem at the DNA Level.Clinical cancer research : an official journal of the American Association for Cancer Research 6/1/2017; 23.11; 2617-2629
[Full Text Article] [Pubmed: 28572256] [PMC: PMC5458139] -
Noh, Songmi et al. Optimal combination of immunohistochemical markers for subclassification of non-small cell lung carcinomas: A tissue microarray study of poorly differentiated areas.Lung Cancer 4/1/2012; 76.1; 51-55
[Full Text Article] [Pubmed: 21978426] -
Oronsky, Bryan et al. Navigating the “No Man's Land” of TKI-Failed EGFR-Mutated Non–Small Cell Lung Cancer (NSCLC): A Review.Neoplasia (New York, N.Y.) 12/8/2017; 20.1; 92-98
[Full Text Article] [Pubmed: 29227909] [PMC: PMC5726883] -
Pruitt, Kim D. et al. NCBI Reference Sequences (RefSeq): current status, new features and genome annotation policy.Nucleic Acids Research 2012-01; 40.Database issue; D130-D135
[Full Text Article] [Pubmed: 22121212] [PMC: PMC3245008] -
Rekhtman, Natasha et al. Immunohistochemical algorithm for differentiation of lung adenocarcinoma and squamous cell carcinoma based on large series of whole-tissue sections with validation in small specimens.Modern Pathology 2011-10; 24.10; 1348
[Full Text Article] -
Rekhtman, Natasha et al. Immunohistochemical algorithm for differentiation of lung adenocarcinoma and squamous cell carcinoma based on large series of whole-tissue sections with validation in small specimens.Modern Pathology 2011-10; 24.10; 1348
[Full Text Article] -
Rekhtman, Natasha et al. Neuroendocrine Tumors of the Lung: An Update.Archives of Pathology & Laboratory Medicine 11/1/2010; 134.11; 1628-1638
[Full Text Article] -
Rothschild, Sacha I. et al. Targeted Therapies in Non-Small Cell Lung Cancer—Beyond EGFR and ALK.Cancers 5/26/2015; 7.2; 930-949
[Full Text Article] [Pubmed: 26018876] [PMC: PMC4491691] -
Rudin, Charles M. et al. Comprehensive genomic analysis identifies SOX2 as a frequently amplified gene in small-cell lung cancer.Nature Genetics 2012-10; 44.10; 1111
[Full Text Article] -
Safe, Stephen et al. Role of the Aryl Hydrocarbon Receptor in Carcinogenesis and Potential as a Drug Target.Toxicological Sciences 9/1/2013; 135.1; 43116
[Full Text Article] -
Sasaki, Hidefumi et al. APOBEC3B gene overexpression in non-small-cell lung cancer.Biomedical Reports 2014-05; 2.3; 392-395
[Full Text Article] [Pubmed: 24748981] [PMC: PMC3990218] -
Savci-Heijink, Cemile Dilara et al. The role of desmoglein-3 in the diagnosis of squamous cell carcinoma of the lung.The American Journal of Pathology 2009-05; 174.5; 1629-1637
[Full Text Article] [Pubmed: 19342368] [PMC: PMC2671252] -
Seregni, E. et al. Clinical significance of blood chromogranin A measurement in neuroendocrine tumours.Annals of Oncology: Official Journal of the European Society for Medical Oncology 2001; 12 Suppl 2; S69-72
[Pubmed: 11762355] -
Shanzhi, Wan et al. The Relationship between TTF-1 Expression and EGFR Mutations in Lung Adenocarcinomas.PLOS ONE 4/17/2014; 9.4; e95479
[Full Text Article] -
Siddiqui, Momin T. et al. TTF-1 and Napsin a Double Staining in Diagnosing Lung Adenocarcinoma.Journal of Cytology & Histology 6/11/2012; 3.3;
[Full Text Article] -
Sorensen, Steffen Filskov et al. PD-L1 Expression and Survival among Patients with Advanced Non–Small Cell Lung Cancer Treated with Chemotherapy.Translational Oncology 2/1/2016; 9.1; 64-69
[Full Text Article] -
Syversen, U. et al. Clinical significance of elevated serum chromogranin A levels.Scandinavian Journal of Gastroenterology 2004-10; 39.10; 969-973
[Full Text Article] [Pubmed: 15513336] -
Takahashi, Hiroki et al. Pulmonary surfactant proteins A and D: innate immune functions and biomarkers for lung diseases.Current Pharmaceutical Design 2006; 12.5; 589-598
[Pubmed: 16472150] -
Takahashi, Kazutoshi et al. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors.Cell 8/25/2006; 126.4; 663-676
[Full Text Article] [Pubmed: 16904174] -
Tan, Dongfeng et al. Immunohistochemistry for Assessment of Pulmonary and Pleural Neoplasms: A Review and Update.International Journal of Clinical and Experimental Pathology 1/1/2008; 1.1; 19-31
[Pubmed: 18784820] [PMC: PMC2480532] -
Taneja, Tarvinder K et al. Markers of small cell lung cancer.World Journal of Surgical Oncology 5/5/2004; 2; 10
[Full Text Article] [Pubmed: 15128454] [PMC: PMC441408] -
Tang, Yanna et al. The association between PD-L1 and EGFR status and the prognostic value of PD-L1 in advanced non-small cell lung cancer patients treated with EGFR-TKIs.Oncotarget 3/29/2015; 6.16; 14209-14219
[Pubmed: 25895031] [PMC: PMC4546461] -
Travis, William D. et al. Update on small cell carcinoma and its differentiation from squamous cell carcinoma and other non-small cell carcinomas.Modern Pathology 1/3/2012; 25.S1; S18
[Full Text Article] -
Tsai, Chi-Hao et al. The inhibition of lung cancer cell migration by AhR-regulated autophagy.Scientific Reports 2/14/2017; 7; 41927
[Full Text Article] -
Tsay, Junchieh J. et al. Aryl Hydrocarbon Receptor and Lung Cancer.Anticancer research 2013-04; 33.4; 1247-1256
[Pubmed: 23564762] [PMC: PMC3771678] -
Turner, Bradley M. et al. Napsin A, a new marker for lung adenocarcinoma, is complementary and more sensitive and specific than thyroid transcription factor 1 in the differential diagnosis of primary pulmonary carcinoma: evaluation of 1674 cases by tissue microarray.Archives of Pathology & Laboratory Medicine 2012-02; 136.2; 163-171
[Full Text Article] [Pubmed: 22288963] -
Ueno, Takayuki et al. Processing of pulmonary surfactant protein B by napsin and cathepsin H.The Journal of Biological Chemistry 4/16/2004; 279.16; 16178-16184
[Full Text Article] [Pubmed: 14766755] -
Van Acker, Heleen H et al. CD56 in the Immune System: More Than a Marker for Cytotoxicity?.Frontiers in Immunology 2017; 8;
[Full Text Article] -
Webb, Thomas R et al. Anaplastic lymphoma kinase: role in cancer pathogenesis and small-molecule inhibitor development for therapy.Expert review of anticancer therapy 2009-03; 9.3; 331-356
[Full Text Article] [Pubmed: 19275511] [PMC: PMC2780428] -
Westcott, Peter M. K. et al. The genetics and biology of KRAS in lung cancer.Chinese Journal of Cancer 2013-02; 32.2; 63-70
[Full Text Article] [Pubmed: 22776234] [PMC: PMC3845615] -
Whyte, Warren A. et al. Master Transcription Factors and Mediator Establish Super-Enhancers at Key Cell Identity Genes.Cell 4/11/2013; 153.2; 307-319
[Full Text Article] [Pubmed: 23582322] -
Wiedenmann, B et al. Synaptophysin: a marker protein for neuroendocrine cells and neoplasms..Proceedings of the National Academy of Sciences of the United States of America 1986-05; 83.10; 3500-3504
[Pubmed: 3010302] [PMC: PMC323544] -
Wu, Wei et al. Non-Canonical Thinking for Targeting ALK-Fusion Onco-Proteins in Lung Cancer.Cancers 11/30/2017; 9.12;
[Full Text Article] [Pubmed: 29189709] [PMC: PMC5742812] -
Yan, Shumei et al. Increased APOBEC3B Predicts Worse Outcomes in Lung Cancer: A Comprehensive Retrospective Study.Journal of Cancer 3/19/2016; 7.6; 618-625
[Full Text Article] [Pubmed: 27076842] [PMC: PMC4829547] -
Ye, Jiqing et al. Combination of napsin A and TTF-1 immunohistochemistry helps in differentiating primary lung adenocarcinoma from metastatic carcinoma in the lung.Applied immunohistochemistry & molecular morphology: AIMM 2011-07; 19.4; 313-317
[Full Text Article] [Pubmed: 21464700] -
Ye, Mingxiang et al. Activation of the Aryl Hydrocarbon Receptor Leads to Resistance to EGFR TKIs in Non-Small-Cell Lung Cancer by Activating Src-Mediated Bypass Signaling.Clinical Cancer Research 1/1/2017; clincanres.0396.2017
[Full Text Article] [Pubmed: 29229632] -
Zhang, Minghui et al. PD-L1 expression in lung cancer and its correlation with driver mutations: a meta-analysis.Scientific Reports 8/31/2017; 7.1; 10255
[Full Text Article] -
Zhu, Qing-Ge et al. Driver genes in non-small cell lung cancer: Characteristics, detection methods, and targeted therapies.Oncotarget 8/22/2017; 8.34; 57680-57692
[Full Text Article] [Pubmed: 28915704] [PMC: PMC5593676]